This page is historical material reflecting the Feedback Loop Blog as it existed on
January 14, 2015. This page is no longer updated and links to external websites
and some internal pages may not work.
January 14, 2015
Researchers have discovered a faster, easier and more affordable technique for processing biological samples. Credit: Weldon School of Biomedical Engineering, Purdue University.
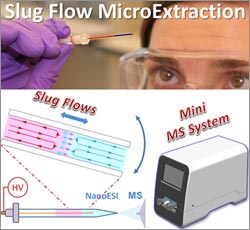
It’s not unusual for the standard dose of a drug to work well for one person but be less effective for another. One reason for such differences is that individuals can break down drugs at different rates, leading to different concentrations of drugs and of their breakdown products (metabolites) in the bloodstream. A promising new process called slug-flow microextraction could make it faster, easier and more affordable to regularly monitor drug metabolites so that medication dosages could be tailored to each patient’s needs, an approach known as personalized medicine. This technique could also allow researchers to better monitor people’s responses to new drug treatments during clinical trials.
Purdue University researchers led by
Zheng Ouyang 
devised the streamlined method, which involves inserting a tiny amount of biological fluid such as blood or urine into a narrow, sealed tube containing an organic solvent. At first the two liquids act like oil and water, but gently rocking the tube back and forth causes small amounts of the drug or other small molecules of interest to move into the solvent. Scientists then can use a large laboratory instrument called a mass spectrometer to analyze the sample in the solvent.
The Purdue team is now developing a desktop mass spectrometer that weighs just 40 pounds, as opposed to the 300-pound conventional device. The goal is to develop a portable technology that can be used by nurses and physicians in community clinics and doctor’s offices—and eventually by patients—in place of analytical labs with bulky and expensive equipment, says Fred Friedman of NIGMS’
Division of Biomedical Technology, Bioinformatics, and Computational Biology. “It’s really a big step forward for personalized medicine.”
This work was funded in part by NIH under grant R01GM106016.