Appropriations Subcommittee Statement on the Fiscal Year 2000 Budget
February 25, 1999
Mr. Chairman and Members of the Committee: The President in his FY 2000 budget has proposed that the National Institute of General Medical Sciences (NIGMS) receive $1.194 billion, an increase of $28 million over the comparable FY 1999 appropriations. Including the estimated allocation for AIDS, the total support proposed for NIGMS is $1.227 billion, an increase of $29 million over the FY 1999 appropriation. Funds for NIGMS efforts in AIDS research are included within the Office of AIDS Research budget request.
I am pleased to present to you the programs of the National Institute of General Medical Sciences. The NIGMS mission is to support basic biomedical research that is not targeted to specific diseases, but that increases understanding of life processes and lays the foundation for advances in disease diagnosis, treatment, and prevention. The Institute also has a major role in training the next generation of scientists. As part of this effort, we attempt to ensure that biomedical research has access to the broadest possible intellectual resources in our society, through programs that provide research and training support for underrepresented minorities.
A Look Back
I would like to begin by describing two important recent research advances that illustrate the long-term nature of the research we support, the way in which it often draws from observations made in a number of different organisms, and the speed with which many of these fundamental studies become relevant to the human condition.
The first advance involves an essential component of the cell, called the microtubule. Microtubules are long, stiff structures that extend through the cell [Figure 1] and are involved in such key functions as cell division and the movement of material within the cell. An understanding of the structure and function of the microtubule has been a major scientific goal for several decades.
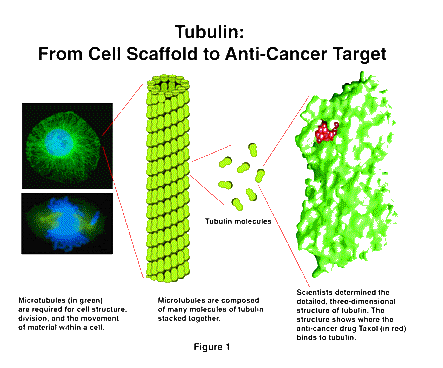
Recently, investigators supported by NIGMS have determined the three-dimensional structure of the units that make up the microtubules, called tubulin. Of particular interest is the fact that the anti-cancer drug, Taxol, acts by binding to tubulin. The location of the Taxol on the tubulin molecule is clearly visible in this structure. The identification of the binding site for Taxol will help in developing new anti-cancer drugs.
This very important structure was determined by a relatively novel technique. Since tubulin spontaneously aggregates into very large sheets, the usual methods for detailed structure determination, X-ray diffraction and nuclear magnetic resonance, could not be used. Instead, the researchers used a powerful variant of electron microscopy, which is increasingly being applied to the analysis of large, complex structures. Development of this tool has been supported by NIGMS for more than two decades, and is now coming to fruition in this and other research areas.
A second major research advance is in the understanding of one of the most pervasive and, until recently, least understood aspects of biological systems, the circadian rhythm. This pattern of activity, with a periodicity of about 24 hours, appears to be present everywhere one looks, from plants to yeast to fruit flies to humans. Disruption of the biological clock is most apparent in the sleep disorders that accompany jet lag, but the clock almost certainly plays a fundamental role in the normal physiology of living organisms. Although studies on the molecular basis of circadian rhythms have been underway for at least 30 years, the last 18 months have yielded an explosion of information on the way that cellular clocks operate. The general mechanism looks quite simple at this point, although the simplicity is undoubtedly deceiving, and much yet remains to be learned [Figure 2]. This simple model shows a feedback loop, where a pair of proteins (the PAS proteins) stimulates the synthesis of the clock proteins. As these increase in concentration, they prevent the PAS proteins from promoting their synthesis, and the concentration of the clock proteins drops. A new cycle is then initiated. The timing of this cycle of synthesis, inhibition, and renewal determines the period of the cellular clock.
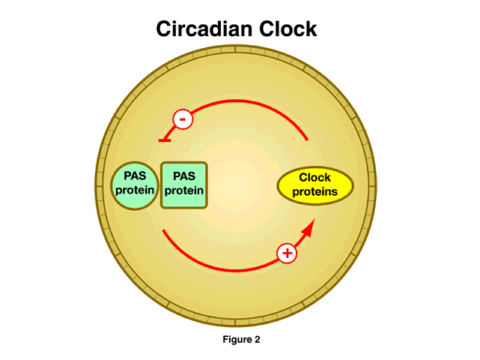
It is striking that very similar proteins exist in all the organisms studied, from yeast to mammals. This similarity also extends to one of the mechanisms by which the cycle is triggered, that is, the response to light. Recently, three NIGMS-supported research teams have identified the way the cells respond to light to modulate this cycle. Again, it is striking that the photoreceptor is the same in the plant model, in fruit flies, and in mice.
A Look Ahead
I would like to spend the rest of my time dealing not with the past, but with the future. However, there are certain common features that have led to the successes of the past, and that we will continue to emphasize in the future. The two examples I just gave demonstrate many of these features. The application of novel technologies, the use of detailed structural information to understand the ways that drugs work, the use of model systems to understand fundamental biological processes, and the application of genomic information to identify proteins with common functions in different organisms, as was done in the studies of biological clocks, are common events in many new discoveries. Another common denominator is the availability of stable, long-term support to allow the resolution of difficult research problems. Finally, these research advances all emerged from peer-reviewed, investigator-initiated, individual research grants.
Recent discussions with advisory groups have also identified a number of new approaches with significant potential payoffs. Most prominently, there was widespread agreement on the need to help support significantly broader collaborative interactions than have been the norm to this time; on the need for access to a broad array of technologies; and on the need for the incorporation into basic research of quantitative disciplines such as mathematics, engineering, physics, and computer science. We have developed, together with our Advisory Council and other groups, an extensive group of initiatives reflecting these needs. Given the time available, I will only discuss two of these in detail.
Voltaire complained that doctors poured drugs of which they knew little to cure diseases of which they knew less into human beings of which they knew nothing. Since then, we have learned a great deal about drugs and diseases, but much less about the humans who are being treated. Our new pharmacogenetics initiative is designed to address this gap in understanding. Pharmacogenetics is the study of differences between individuals in the response to drugs, using the tools of genetics.
An example of what is involved is shown in the next figure [Figure 3]. This is the result of a study by an NIGMS investigator showing that the response to an anti-leukemia drug can vary significantly among the treated population. The drug is not only therapeutic, but it can be toxic if it remains in the system too long. In most people, it is rapidly degraded, and the doses are balanced to provide the maximum benefit and the minimum toxicity. However, in a small number of individuals the drug is very poorly degraded, and the results can be fatal. The study showed that the differences in response came from the variation in a gene for a specific enzyme that is involved in the degradation of the drug. Because this is now understood, a simple blood test can determine the appropriate drug levels for this treatment.
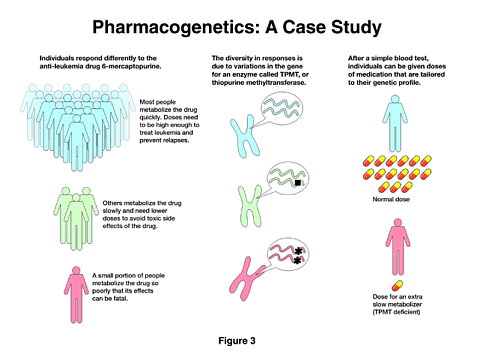
We would like to expand our ability to identify such differences between individuals and thus provide the most appropriate treatments. Consequently, we are planning to support the development of a network of multidisciplinary research groups to identify the functional variations in genes and enzymes that determine drug responses. At the same time, we will create a pharmacogenetic database in which to store, analyze, and access the information for future applications. As I noted above, access to research tools is essential for further progress, and we believe that the database I have described will be an important tool for pharmacologists and scientists generally.
The second initiative I want to describe builds on the extraordinary possibilities presented to us by the complete understanding of genomes, both the human genome and those of other organisms. Our goal at NIGMS is to arrive at a complete understanding of how cells function. Knowledge of the genes is the indispensable starting point, since they determine and regulate the production of the proteins that conduct the cell's business. The next step is to understand how these proteins function, and, as I demonstrated in the example of tubulin, this is tightly linked to an understanding of structure. As shown in the next figure [Figure 4], we propose to systematically analyze families of proteins to get a reasonably complete catalog of all the representative protein structures. We expect this to provide many benefits for investigators who are conducting research on the relationship of protein structure to function, including an understanding of the way aberrant proteins result in disease.
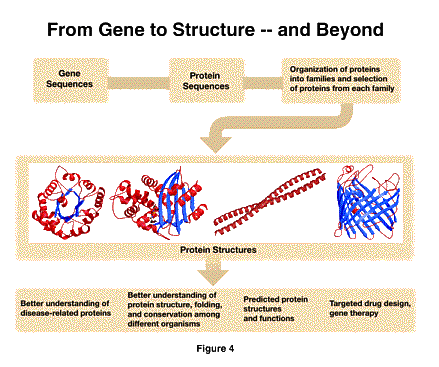
This initiative is the result of workshops and planning meetings over more than a year, involving several agencies (most notably the Department of Energy) and representatives of the scientific community, including scientists from both Europe and Japan. We expect to develop this effort as a close inter-agency and international collaboration.
Last, but hardly least, the evolution of the biological sciences continues to require the incorporation of new skills in the training of investigators. We have initiated new programs to bring into biology investigators with training in quantitative disciplines; to provide support for outstanding physician-scientists to be trained in research in the areas of anesthesiology, clinical pharmacology, and trauma and burn injury; and to help postdoctoral trainees improve their teaching skills by combining a traditional research experience with mentored teaching at a minority-serving institution. We expect these and other initiatives to greatly improve and expand the capabilities of our researchers, to develop new areas of science, to broaden and enhance training opportunities, and to stimulate the entry of underrepresented minorities into basic biomedical research.
The activities of NIGMS are covered within the NIH-wide Annual Performance Plan required under the Government Performance and Results Act (GPRA). The FY 2000 performance goals and measures for NIH are detailed in this performance plan and are linked to both the budget and the HHS GPRA Strategic Plan, which was transmitted to Congress on September 30, 1997. NIH's performance targets in the Plan are partially a function of resource levels requested in the President's Budget and could change based upon final Congressional Appropriations action. NIH looks forward to Congress' feedback on the usefulness of its Performance Plan, as well as to working with Congress on achieving the NIH goals laid out in this plan.
Thank you, Mr. Chairman. I would be pleased to answer any questions that you may have.
