Switch to List View
Image and Video Gallery
This is a searchable collection of scientific photos, illustrations, and videos. The images and videos in this gallery are licensed under Creative Commons Attribution Non-Commercial ShareAlike 3.0. This license lets you remix, tweak, and build upon this work non-commercially, as long as you credit and license your new creations under identical terms.
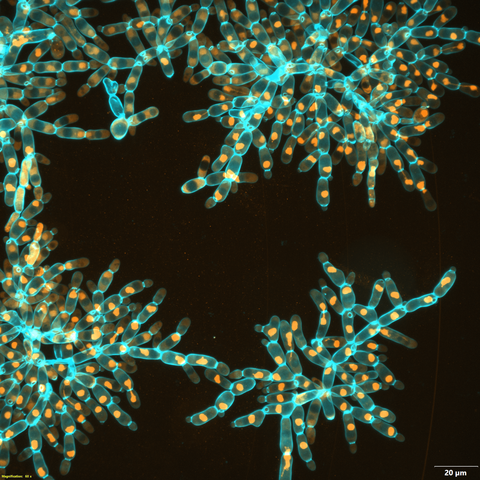
6971: Snowflake yeast 3
6971: Snowflake yeast 3
Multicellular yeast called snowflake yeast that researchers created through many generations of directed evolution from unicellular yeast. Here, the researchers visualized nuclei in orange to help them study changes in how the yeast cells divided. Cell walls are shown in blue. This image was captured using spinning disk confocal microscopy.
Related to images 6969 and 6970.
Related to images 6969 and 6970.
William Ratcliff, Georgia Institute of Technology.
View Media
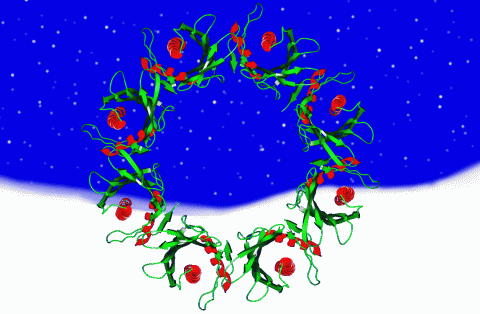
2372: Wreath-shaped protein from X. campestris
2372: Wreath-shaped protein from X. campestris
Crystal structure of a protein with unknown function from Xanthomonas campestris, a plant pathogen. Eight copies of the protein crystallized to form a ring. Chosen as the December 2007 Protein Structure Initiative Structure of the Month.
Ken Schwinn and Sonia Espejon-Reynes, New York SGX Research Center for Structural Genomics
View Media
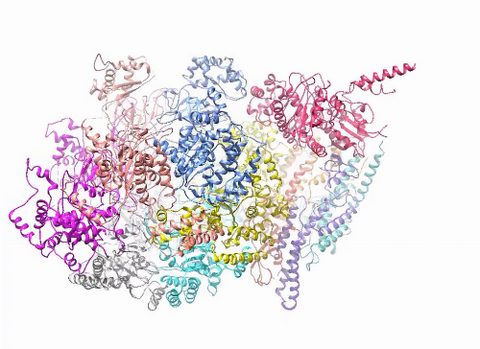
3750: A dynamic model of the DNA helicase protein complex
3750: A dynamic model of the DNA helicase protein complex
This short video shows a model of the DNA helicase in yeast. This DNA helicase has 11 proteins that work together to unwind DNA during the process of copying it, called DNA replication. Scientists used a technique called cryo-electron microscopy (cryo-EM), which allowed them to study the helicase structure in solution rather than in static crystals. Cryo-EM in combination with computer modeling therefore allows researchers to see movements and other dynamic changes in the protein. The cryo-EM approach revealed the helicase structure at much greater resolution than could be obtained before. The researchers think that a repeated motion within the protein as shown in the video helps it move along the DNA strand. To read more about DNA helicase and this proposed mechanism, see this news release by Brookhaven National Laboratory.
Huilin Li, Stony Brook University
View Media
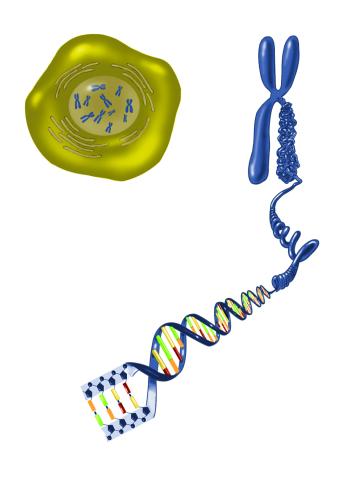
2539: Chromosome inside nucleus
2539: Chromosome inside nucleus
The long, stringy DNA that makes up genes is spooled within chromosomes inside the nucleus of a cell. (Note that a gene would actually be a much longer stretch of DNA than what is shown here.) See image 2540 for a labeled version of this illustration. Featured in The New Genetics.
Crabtree + Company
View Media
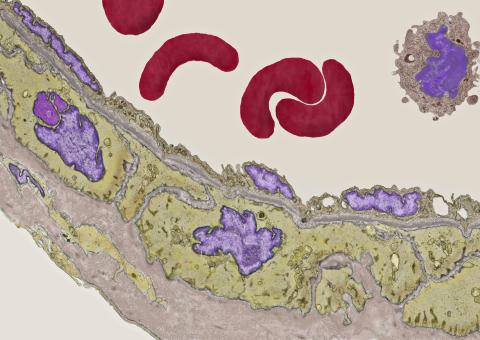
3738: Transmission electron microscopy of coronary artery wall with elastin-rich ECM pseudocolored in light brown
3738: Transmission electron microscopy of coronary artery wall with elastin-rich ECM pseudocolored in light brown
Elastin is a fibrous protein in the extracellular matrix (ECM). It is abundant in artery walls like the one shown here. As its name indicates, elastin confers elasticity. Elastin fibers are at least five times stretchier than rubber bands of the same size. Tissues that expand, such as blood vessels and lungs, need to be both strong and elastic, so they contain both collagen (another ECM protein) and elastin. In this photo, the elastin-rich ECM is colored grayish brown and is most visible at the bottom of the photo. The curved red structures near the top of the image are red blood cells.
Tom Deerinck, National Center for Microscopy and Imaging Research (NCMIR)
View Media
2763: Fused, dicentric chromosomes
2763: Fused, dicentric chromosomes
This fused chromosome has two functional centromeres, shown as two sets of red and green dots. Centromeres are DNA/protein complexes that are key to splitting the chromosomes evenly during cell division. When dicentric chromosomes like this one are formed in a person, fertility problems or other difficulties may arise. Normal chromosomes carrying a single centromere (one set of red and green dots) are also visible in this image.
Beth A. Sullivan, Duke University
View Media

1287: Mitochondria
1287: Mitochondria
Bean-shaped mitochondria are cells' power plants. These organelles have their own DNA and replicate independently. The highly folded inner membranes are the site of energy generation.
Judith Stoffer
View Media
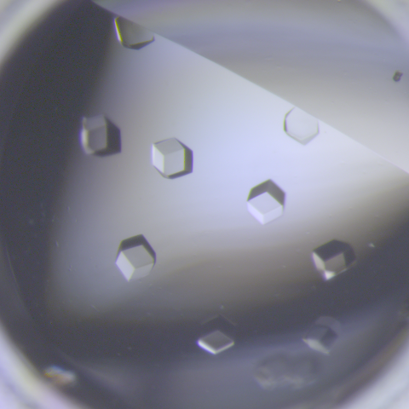
6764: Crystals of CCD-1 in complex with cefotaxime
6764: Crystals of CCD-1 in complex with cefotaxime
CCD-1 is an enzyme produced by the bacterium Clostridioides difficile that helps it resist antibiotics. Here, researchers crystallized bound pairs of CCD-1 molecules and molecules of the antibiotic cefotaxime. This enabled their structure to be studied using X-ray crystallography.
Related to images 6765, 6766, and 6767.
Related to images 6765, 6766, and 6767.
Keith Hodgson, Stanford University.
View Media
6597: Pathways – Bacteria vs. Viruses: What's the Difference?
6597: Pathways – Bacteria vs. Viruses: What's the Difference?
Learn about how bacteria and viruses differ, how they each can make you sick, and how they can or cannot be treated. Discover more resources from NIGMS’ Pathways collaboration with Scholastic. View the video on YouTube for closed captioning.
National Institute of General Medical Sciences
View Media
6520: HeLa cell undergoing division into two daughter cells
6520: HeLa cell undergoing division into two daughter cells
Here, a human HeLa cell (a type of immortal cell line used in laboratory experiments) is undergoing cell division. They come from cervical cancer cells that were obtained in 1951 from Henrietta Lacks, a patient at the Johns Hopkins Hospital. The final stage of division, called cytokinesis, occurs after the genomes—shown in yellow—have split into two new daughter cells. The myosin II is a motor protein shown in blue, and the actin filaments, which are types of protein that support cell structure, are shown in red.
Dylan T. Burnette, Ph.D., Vanderbilt University School of Medicine.
View Media
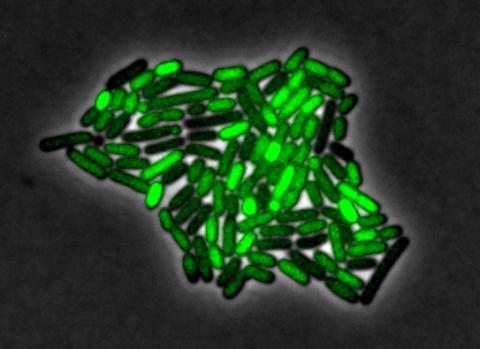
3253: Pulsating response to stress in bacteria
3253: Pulsating response to stress in bacteria
By attaching fluorescent proteins to the genetic circuit responsible for B. subtilis's stress response, researchers can observe the cells' pulses as green flashes. In response to a stressful environment like one lacking food, B. subtilis activates a large set of genes that help it respond to the hardship. Instead of leaving those genes on as previously thought, researchers discovered that the bacteria flip the genes on and off, increasing the frequency of these pulses with increasing stress. See entry 3254 for the related video.
Michael Elowitz, Caltech University
View Media
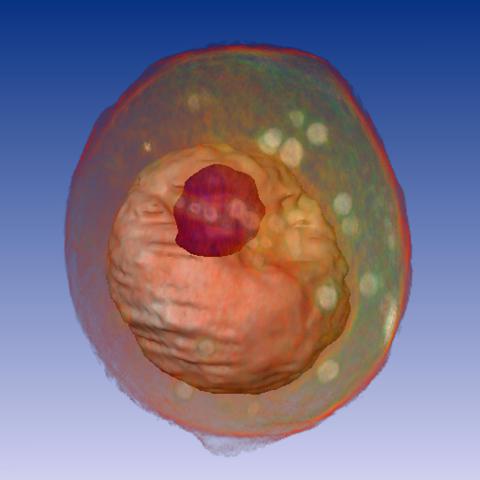
1092: Yeast cell
1092: Yeast cell
A whole yeast (Saccharomyces cerevisiae) cell viewed by X-ray microscopy. Inside, the nucleus and a large vacuole (red) are visible.
Carolyn Larabell, University of California, San Francisco and the Lawrence Berkeley National Laboratory
View Media
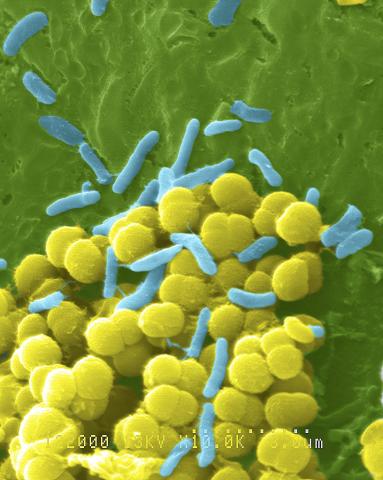
1158: Bacteria shapes
1158: Bacteria shapes
A colorized scanning electron micrograph of bacteria. Scanning electron microscopes allow scientists to see the three-dimensional surface of their samples.
Tina Weatherby Carvalho, University of Hawaii at Manoa
View Media
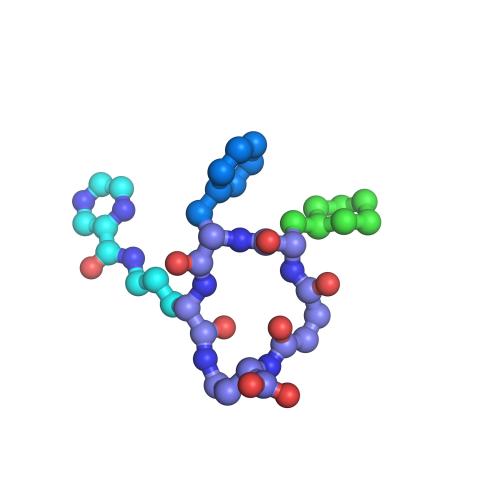
3419: X-ray co-crystal structure of Src kinase bound to a DNA-templated macrocycle inhibitor 7
3419: X-ray co-crystal structure of Src kinase bound to a DNA-templated macrocycle inhibitor 7
X-ray co-crystal structure of Src kinase bound to a DNA-templated macrocycle inhibitor. Related to images 3413, 3414, 3415, 3416, 3417, and 3418.
Markus A. Seeliger, Stony Brook University Medical School and David R. Liu, Harvard University
View Media
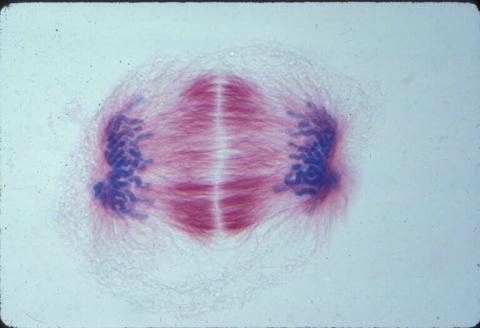
1018: Lily mitosis 12
1018: Lily mitosis 12
A light microscope image of a cell from the endosperm of an African globe lily (Scadoxus katherinae). This is one frame of a time-lapse sequence that shows cell division in action. The lily is considered a good organism for studying cell division because its chromosomes are much thicker and easier to see than human ones. Staining shows microtubules in red and chromosomes in blue. Here, condensed chromosomes are clearly visible near the end of a round of mitosis.
Related to images 1010, 1011, 1012, 1013, 1014, 1015, 1016, 1017, 1019, and 1021.
Related to images 1010, 1011, 1012, 1013, 1014, 1015, 1016, 1017, 1019, and 1021.
Andrew S. Bajer, University of Oregon, Eugene
View Media

2724: Blinking bacteria
2724: Blinking bacteria
Like a pulsing blue shower, E. coli cells flash in synchrony. Genes inserted into each cell turn a fluorescent protein on and off at regular intervals. When enough cells grow in the colony, a phenomenon called quorum sensing allows them to switch from blinking independently to blinking in unison. Researchers can watch waves of light propagate across the colony. Adjusting the temperature, chemical composition or other conditions can change the frequency and amplitude of the waves. Because the blinks react to subtle changes in the environment, synchronized oscillators like this one could one day allow biologists to build cellular sensors that detect pollutants or help deliver drugs.
Jeff Hasty, University of California, San Diego
View Media
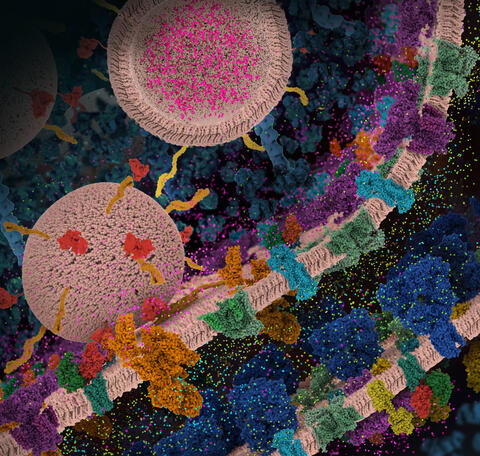
6992: Molecular view of glutamatergic synapse
6992: Molecular view of glutamatergic synapse
This illustration highlights spherical pre-synaptic vesicles that carry the neurotransmitter glutamate. The presynaptic and postsynaptic membranes are shown with proteins relevant for transmitting and modulating the neuronal signal.
PDB 101’s Opioids and Pain Signaling video explains how glutamatergic synapses are involved in the process of pain signaling.
PDB 101’s Opioids and Pain Signaling video explains how glutamatergic synapses are involved in the process of pain signaling.
Amy Wu and Christine Zardecki, RCSB Protein Data Bank.
View Media
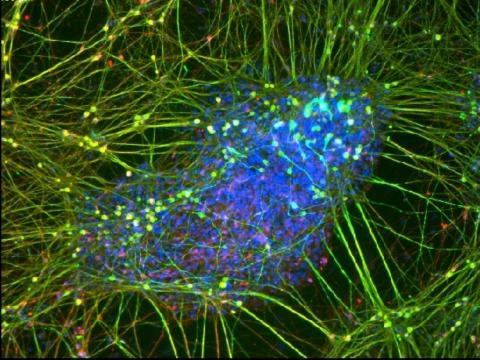
3264: Peripheral nerve cell derived from ES cells
3264: Peripheral nerve cell derived from ES cells
A peripheral nerve cell made from human embryonic stem cell-derived neural crest stem cells. The nucleus is shown in blue, and nerve cell proteins peripherin and beta-tubulin (Tuj1) are shown in green and red, respectively. Related to image 3263.
Stephen Dalton, University of Georgia
View Media

3719: CRISPR illustration
3719: CRISPR illustration
This illustration shows, in simplified terms, how the CRISPR-Cas9 system can be used as a gene-editing tool.
For an explanation and overview of the CRISPR-Cas9 system, see the iBiology video, and download the four images of the CRIPSR illustration here.
For an explanation and overview of the CRISPR-Cas9 system, see the iBiology video, and download the four images of the CRIPSR illustration here.
National Institute of General Medical Sciences.
View Media

2398: RNase A (1)
2398: RNase A (1)
A crystal of RNase A protein created for X-ray crystallography, which can reveal detailed, three-dimensional protein structures.
Alex McPherson, University of California, Irvine
View Media

6902: Arachnoidiscus diatom
6902: Arachnoidiscus diatom
An Arachnoidiscus diatom with a diameter of 190µm. Diatoms are microscopic algae that have cell walls made of silica, which is the strongest known biological material relative to its density. In Arachnoidiscus, the cell wall is a radially symmetric pillbox-like shell composed of overlapping halves that contain intricate and delicate patterns. Sometimes, Arachnoidiscus is called “a wheel of glass.”
This image was taken with the orientation-independent differential interference contrast microscope.
This image was taken with the orientation-independent differential interference contrast microscope.
Michael Shribak, Marine Biological Laboratory/University of Chicago.
View Media
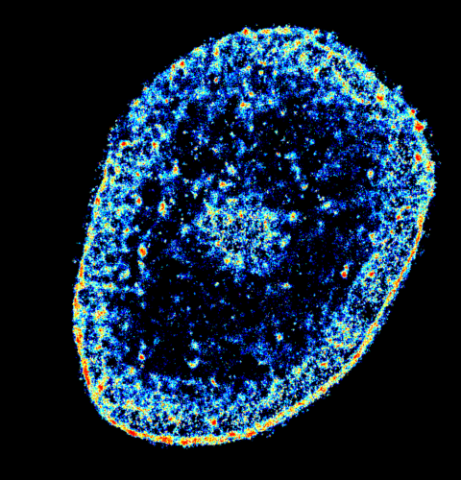
6893: Chromatin in human tenocyte
6893: Chromatin in human tenocyte
The nucleus of a degenerating human tendon cell, also known as a tenocyte. It has been color-coded based on the density of chromatin—a substance made up of DNA and proteins. Areas of low chromatin density are shown in blue, and areas of high chromatin density are shown in red. This image was captured using Stochastic Optical Reconstruction Microscopy (STORM).
Related to images 6887 and 6888.
Related to images 6887 and 6888.
Melike Lakadamyali, Perelman School of Medicine at the University of Pennsylvania.
View Media
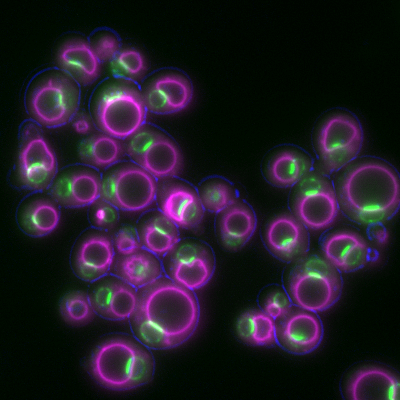
6772: Yeast cells responding to a glucose shortage
6772: Yeast cells responding to a glucose shortage
These yeast cells were exposed to a glucose (sugar) shortage. This caused the cells to compartmentalize HMGCR (green)—an enzyme involved in making cholesterol—to a patch on the nuclear envelope next to the vacuole/lysosome (purple). This process enhanced HMGCR activity and helped the yeast adapt to the glucose shortage. Researchers hope that understanding how yeast regulate cholesterol could ultimately lead to new ways to treat high cholesterol in people. This image was captured using a fluorescence microscope.
Mike Henne, University of Texas Southwestern Medical Center.
View Media
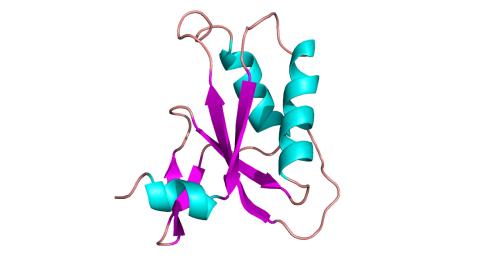
3428: Antitoxin GhoS (Illustration 2)
3428: Antitoxin GhoS (Illustration 2)
Structure of the bacterial antitoxin protein GhoS. GhoS inhibits the production of a bacterial toxin, GhoT, which can contribute to antibiotic resistance. GhoS is the first known bacterial antitoxin that works by cleaving the messenger RNA that carries the instructions for making the toxin. More information can be found in the paper: Wang X, Lord DM, Cheng HY, Osbourne DO, Hong SH, Sanchez-Torres V, Quiroga C, Zheng K, Herrmann T, Peti W, Benedik MJ, Page R, Wood TK. A new type V toxin-antitoxin system where mRNA for toxin GhoT is cleaved by antitoxin GhoS. Nat Chem Biol. 2012 Oct;8(10):855-61. Related to 3427.
Rebecca Page and Wolfgang Peti, Brown University and Thomas K. Wood, Pennsylvania State University
View Media

3733: A molecular interaction network in yeast 3
3733: A molecular interaction network in yeast 3
The image visualizes a part of the yeast molecular interaction network. The lines in the network represent connections among genes (shown as little dots) and different-colored networks indicate subnetworks, for instance, those in specific locations or pathways in the cell. Researchers use gene or protein expression data to build these networks; the network shown here was visualized with a program called Cytoscape. By following changes in the architectures of these networks in response to altered environmental conditions, scientists can home in on those genes that become central "hubs" (highly connected genes), for example, when a cell encounters stress. They can then further investigate the precise role of these genes to uncover how a cell's molecular machinery deals with stress or other factors. Related to images 3730 and 3732.
Keiichiro Ono, UCSD
View Media
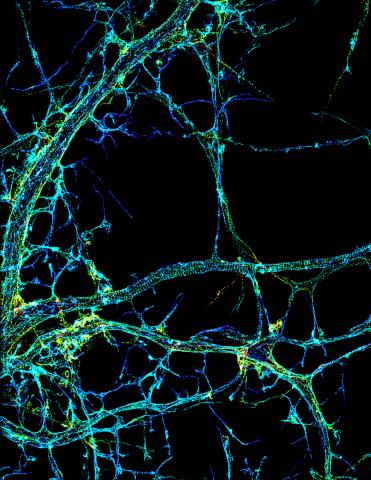
3678: STORM image of axonal cytoskeleton
3678: STORM image of axonal cytoskeleton
This image shows the long, branched structures (axons) of nerve cells. Running horizontally across the middle of the photo is an axon wrapped in rings made of actin protein (green), which plays important roles in nerve cells. The image was captured with a powerful microscopy technique that allows scientists to see single molecules in living cells in real time. The technique is called stochastic optical reconstruction microscopy (STORM). It is based on technology so revolutionary that its developers earned the 2014 Nobel Prize in Chemistry. More information about this image can be found in: K. Xu, G. Zhong, X. Zhuang. Actin, spectrin and associated proteins form a periodic cytoskeleton structure in axons. Science 339, 452-456 (2013).
Xiaowei Zhuang Laboratory, Howard Hughes Medical Institute, Harvard University
View Media

5872: Mouse retina close-up
5872: Mouse retina close-up
Keunyoung ("Christine") Kim National Center for Microscopy and Imaging Research (NCMIR)
View Media
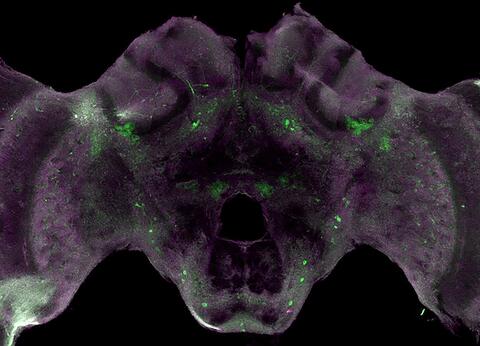
6755: Honeybee brain
6755: Honeybee brain
Insect brains, like the honeybee brain shown here, are very different in shape from human brains. Despite that, bee and human brains have a lot in common, including many of the genes and neurochemicals they rely on in order to function. The bright-green spots in this image indicate the presence of tyrosine hydroxylase, an enzyme that allows the brain to produce dopamine. Dopamine is involved in many important functions, such as the ability to experience pleasure. This image was captured using confocal microscopy.
Gene Robinson, University of Illinois at Urbana-Champaign.
View Media
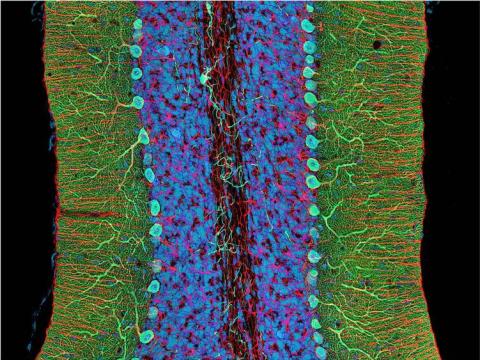
3371: Mouse cerebellum close-up
3371: Mouse cerebellum close-up
The cerebellum is the brain's locomotion control center. Every time you shoot a basketball, tie your shoe or chop an onion, your cerebellum fires into action. Found at the base of your brain, the cerebellum is a single layer of tissue with deep folds like an accordion. People with damage to this region of the brain often have difficulty with balance, coordination and fine motor skills. For a lower magnification, see image 3639.
This image was part of the Life: Magnified exhibit that ran from June 3, 2014, to January 21, 2015, at Dulles International Airport.
This image was part of the Life: Magnified exhibit that ran from June 3, 2014, to January 21, 2015, at Dulles International Airport.
National Center for Microscopy and Imaging Research (NCMIR)
View Media
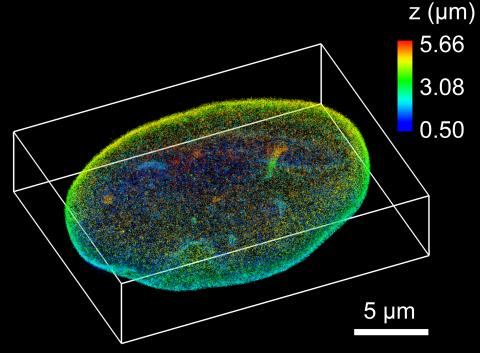
6572: Nuclear Lamina
6572: Nuclear Lamina
The 3D single-molecule super-resolution reconstruction of the entire nuclear lamina in a HeLa cell was acquired using the TILT3D platform. TILT3D combines a tilted light sheet with point-spread function (PSF) engineering to provide a flexible imaging platform for 3D single-molecule super-resolution imaging in mammalian cells.
See 6573 for 3 separate views of this structure.
See 6573 for 3 separate views of this structure.
Anna-Karin Gustavsson, Ph.D.
View Media
2475: Chromosome fiber 01
2475: Chromosome fiber 01
This microscopic image shows a chromatin fiber--a DNA molecule bound to naturally occurring proteins.
Marc Green and Susan Forsburg, University of Southern California
View Media
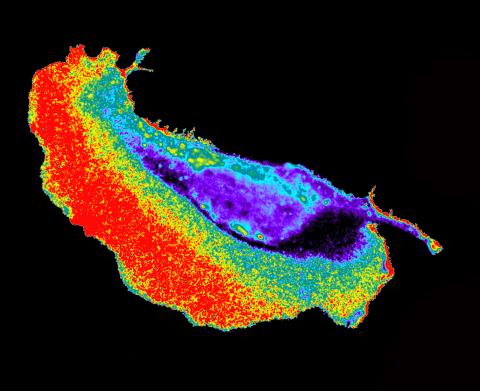
2452: Seeing signaling protein activation in cells 02
2452: Seeing signaling protein activation in cells 02
Cdc42, a member of the Rho family of small guanosine triphosphatase (GTPase) proteins, regulates multiple cell functions, including motility, proliferation, apoptosis, and cell morphology. In order to fulfill these diverse roles, the timing and location of Cdc42 activation must be tightly controlled. Klaus Hahn and his research group use special dyes designed to report protein conformational changes and interactions, here in living neutrophil cells. Warmer colors in this image indicate higher levels of activation. Cdc42 looks to be activated at cell protrusions.
Related to images 2451, 2453, and 2454.
Related to images 2451, 2453, and 2454.
Klaus Hahn, University of North Carolina, Chapel Hill Medical School
View Media
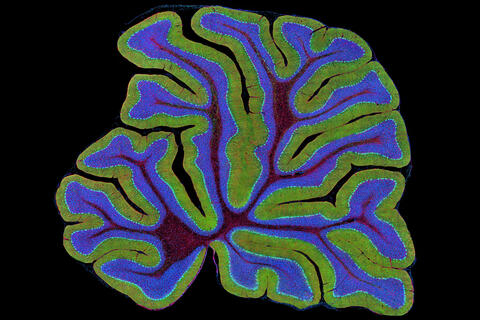
3639: Cerebellum: the brain's locomotion control center
3639: Cerebellum: the brain's locomotion control center
The cerebellum of a mouse is shown here in cross-section. The cerebellum is the brain's locomotion control center. Every time you shoot a basketball, tie your shoe or chop an onion, your cerebellum fires into action. Found at the base of your brain, the cerebellum is a single layer of tissue with deep folds like an accordion. People with damage to this region of the brain often have difficulty with balance, coordination and fine motor skills. For a higher magnification, see image 3371.
This image was part of the Life: Magnified exhibit that ran from June 3, 2014, to January 21, 2015, at Dulles International Airport.
This image was part of the Life: Magnified exhibit that ran from June 3, 2014, to January 21, 2015, at Dulles International Airport.
Thomas Deerinck, National Center for Microscopy and Imaging Research, University of California, San Diego
View Media
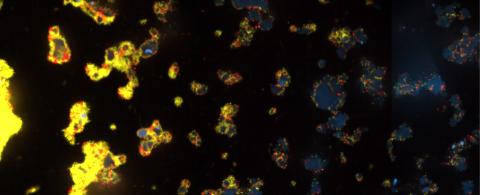
3580: V. Cholerae Biofilm
3580: V. Cholerae Biofilm
Industrious V. cholerae bacteria (yellow) tend to thrive in denser biofilms (left) while moochers (red) thrive in weaker biofilms (right). More information about the research behind this image can be found in a Biomedical Beat Blog posting from February 2014.
View Media
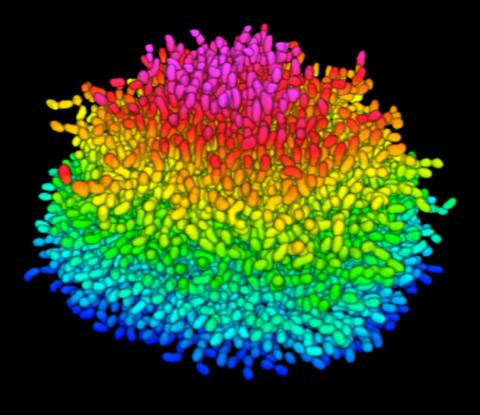
5825: A Growing Bacterial Biofilm
5825: A Growing Bacterial Biofilm
A growing Vibrio cholerae (cholera) biofilm. Cholera bacteria form colonies called biofilms that enable them to resist antibiotic therapy within the body and other challenges to their growth.
Each slightly curved comma shape represents an individual bacterium from assembled confocal microscopy images. Different colors show each bacterium’s position in the biofilm in relation to the surface on which the film is growing.
Each slightly curved comma shape represents an individual bacterium from assembled confocal microscopy images. Different colors show each bacterium’s position in the biofilm in relation to the surface on which the film is growing.
Jing Yan, Ph.D., and Bonnie Bassler, Ph.D., Department of Molecular Biology, Princeton University, Princeton, NJ.
View Media
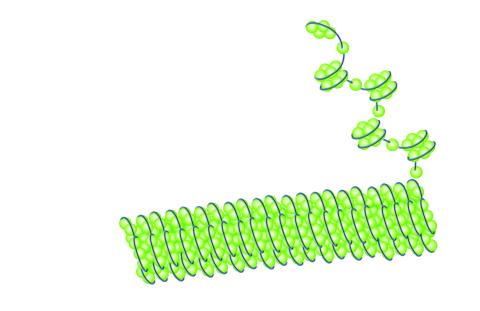
2560: Histones in chromatin
2560: Histones in chromatin
Histone proteins loop together with double-stranded DNA to form a structure that resembles beads on a string. See image 2561 for a labeled version of this illustration. Featured in The New Genetics.
Crabtree + Company
View Media
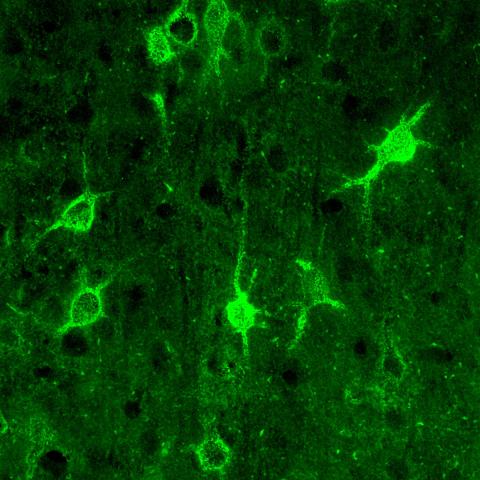
3741: Confocal microscopy of perineuronal nets in the brain 1
3741: Confocal microscopy of perineuronal nets in the brain 1
The photo shows a confocal microscopy image of perineuronal nets (PNNs), which are specialized extracellular matrix (ECM) structures in the brain. The PNN surrounds some nerve cells in brain regions including the cortex, hippocampus and thalamus. Researchers study the PNN to investigate their involvement stabilizing the extracellular environment and forming nets around nerve cells and synapses in the brain. Abnormalities in the PNNs have been linked to a variety of disorders, including epilepsy and schizophrenia, and they limit a process called neural plasticity in which new nerve connections are formed. To visualize the PNNs, researchers labeled them with Wisteria floribunda agglutinin (WFA)-fluorescein. Related to image 3742.
Tom Deerinck, National Center for Microscopy and Imaging Research (NCMIR)
View Media
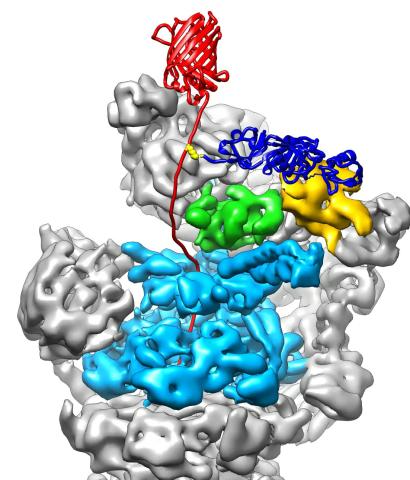
3763: The 26S proteasome engages with a protein substrate
3763: The 26S proteasome engages with a protein substrate
The proteasome is a critical multiprotein complex in the cell that breaks down and recycles proteins that have become damaged or are no longer needed. This illustration shows a protein substrate (red) that is bound through its ubiquitin chain (blue) to one of the ubiquitin receptors of the proteasome (Rpn10, yellow). The substrate's flexible engagement region gets engaged by the AAA+ motor of the proteasome (cyan), which initiates mechanical pulling, unfolding and movement of the protein into the proteasome's interior for cleavage into small shorter protein pieces called peptides. During movement of the substrate, its ubiquitin modification gets cleaved off by the deubiquitinase Rpn11 (green), which sits directly above the entrance to the AAA+ motor pore and acts as a gatekeeper to ensure efficient ubiquitin removal, a prerequisite for fast protein breakdown by the 26S proteasome. Related to video 3764.
Andreas Martin, HHMI
View Media
2536: G switch
2536: G switch
The G switch allows our bodies to respond rapidly to hormones. See images 2537 and 2538 for labeled versions of this image. Featured in Medicines By Design.
Crabtree + Company
View Media
6983: Genetic mosaicism in fruit flies
6983: Genetic mosaicism in fruit flies
Fat tissue from the abdomen of a genetically mosaic adult fruit fly. Genetic mosaicism means that the fly has cells with different genotypes even though it formed from a single zygote. This specific mosaicism results in accumulation of a critical fly adipokine (blue-green) within the fat tissue cells that have reduced expression a key nutrient sensing gene (in left panel). The dotted line shows the cells lacking the gene that is present and functioning in the rest of the cells. Nuclei are labelled in magenta. This image was captured using a confocal microscope and shows a maximum intensity projection of many slices.
Related to images 6982, 6984, and 6985.
Related to images 6982, 6984, and 6985.
Akhila Rajan, Fred Hutchinson Cancer Center
View Media
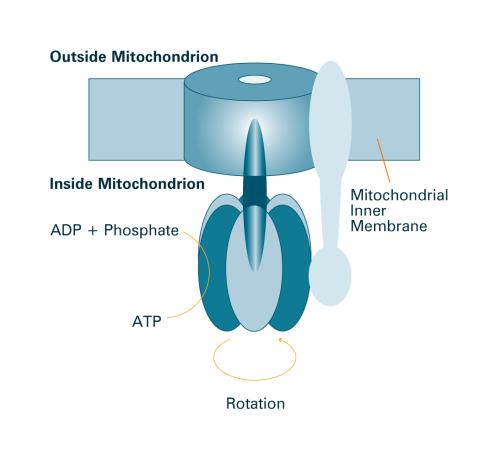
2518: ATP synthase (with labels)
2518: ATP synthase (with labels)
The world's smallest motor, ATP synthase, generates energy for the cell. See image 2517 for an unlabeled version of this illustration. Featured in The Chemistry of Health.
Crabtree + Company
View Media
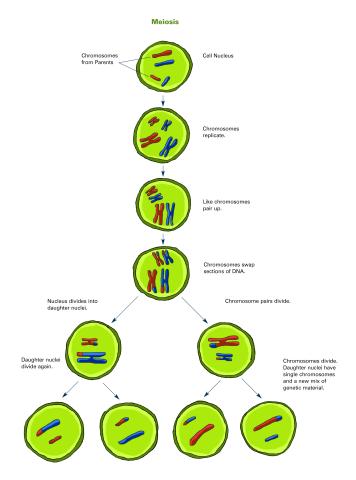
2546: Meiosis illustration (with labels)
2546: Meiosis illustration (with labels)
Meiosis is the process whereby a cell reduces its chromosomes from diploid to haploid in creating eggs or sperm. See image 2545 for an unlabeled version of this illustration. See image 2544 for an unlabeled version of this illustration. Featured in The New Genetics.
Crabtree + Company
View Media
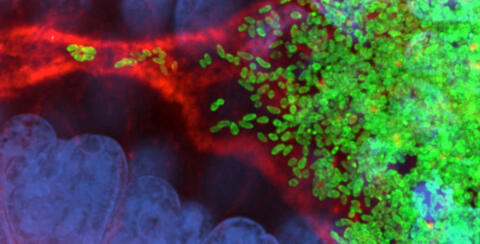
7015: Bacterial cells migrating through the tissues of the squid light organ
7015: Bacterial cells migrating through the tissues of the squid light organ
Vibrio fischeri cells (~ 2 mm), labeled with green fluorescent protein (GFP), passing through a very narrow bottleneck in the tissues (red) of the Hawaiian bobtail squid, Euprymna scolopes, on the way to the crypts where the symbiont population resides. This image was taken using a confocal fluorescence microscope.
Margaret J. McFall-Ngai, Carnegie Institution for Science/California Institute of Technology, and Edward G. Ruby, California Institute of Technology.
View Media
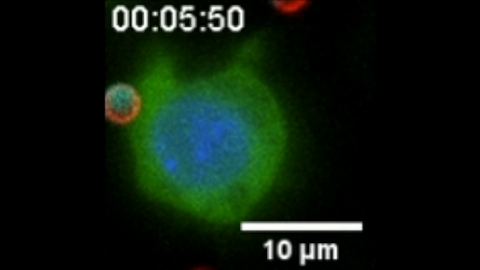
6801: “Two-faced” Janus particle activating a macrophage
6801: “Two-faced” Janus particle activating a macrophage
A macrophage—a type of immune cell that engulfs invaders—“eats” and is activated by a “two-faced” Janus particle. The particle is called “two-faced” because each of its two hemispheres is coated with a different type of molecule, shown here in red and cyan. During macrophage activation, a transcription factor tagged with a green fluorescence protein (NF-κB) gradually moves from the cell’s cytoplasm into its nucleus and causes DNA transcription. The distribution of molecules on “two-faced” Janus particles can be altered to control the activation of immune cells. Details on this “geometric manipulation” strategy can be found in the Proceedings of the National Academy of Sciences paper "Geometrical reorganization of Dectin-1 and TLR2 on single phagosomes alters their synergistic immune signaling" by Li et al. and the Scientific Reports paper "Spatial organization of FcγR and TLR2/1 on phagosome membranes differentially regulates their synergistic and inhibitory receptor crosstalk" by Li et al. This video was captured using epi-fluorescence microscopy.
Related to video 6800.
Related to video 6800.
Yan Yu, Indiana University, Bloomington.
View Media
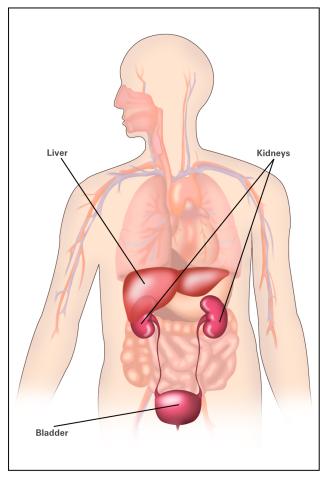
2497: Body toxins (with labels)
2497: Body toxins (with labels)
Body organs such as the liver and kidneys process chemicals and toxins. These "target" organs are susceptible to damage caused by these substances. See image 2496 for an unlabeled version of this illustration.
Crabtree + Company
View Media
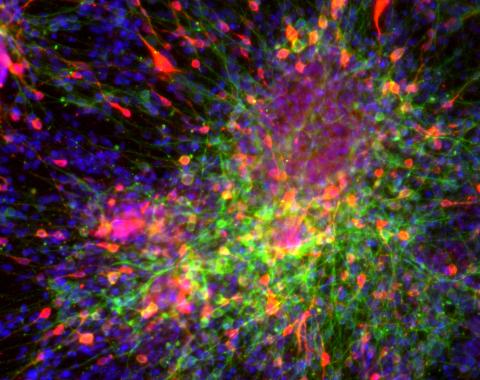
3271: Dopaminergic neurons derived from mouse embryonic stem cells
3271: Dopaminergic neurons derived from mouse embryonic stem cells
These neurons are derived from mouse embryonic stem cells. Red shows cells making a protein called TH that is characteristic of the neurons that degenerate in Parkinson's disease. Green indicates a protein that's found in all neurons. Blue indicates the nuclei of all cells. Studying dopaminergic neurons can help researchers understand the origins of Parkinson's disease and could be used to screen potential new drugs. Image and caption information courtesy of the California Institute for Regenerative Medicine. Related to images 3270 and 3285.
Yaping Sun, lab of Su Guo, University of California, San Francisco, via CIRM
View Media
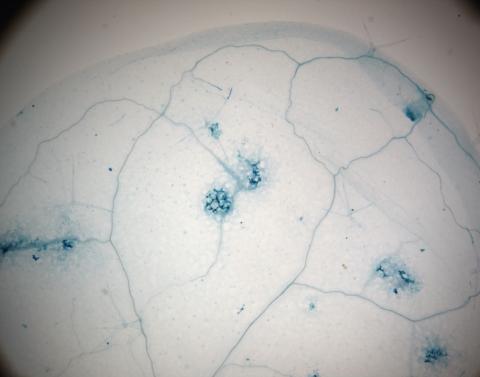
2781: Disease-resistant Arabidopsis leaf
2781: Disease-resistant Arabidopsis leaf
This is a magnified view of an Arabidopsis thaliana leaf a few days after being exposed to the pathogen Hyaloperonospora arabidopsidis. The plant from which this leaf was taken is genetically resistant to the pathogen. The spots in blue show areas of localized cell death where infection occurred, but it did not spread. Compare this response to that shown in Image 2782. Jeff Dangl has been funded by NIGMS to study the interactions between pathogens and hosts that allow or suppress infection.
Jeff Dangl, University of North Carolina, Chapel Hill
View Media
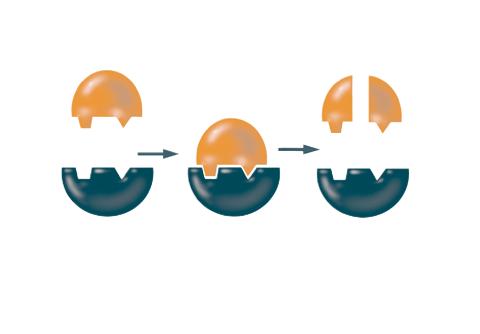
2521: Enzymes convert subtrates into products
2521: Enzymes convert subtrates into products
Enzymes convert substrates into products very quickly. See image 2522 for a labeled version of this illustration. Featured in The Chemistry of Health.
Crabtree + Company
View Media

6804: Staphylococcus aureus in the porous coating of a femoral hip stem
6804: Staphylococcus aureus in the porous coating of a femoral hip stem
Staphylococcus aureus bacteria (blue) on the porous coating of a femoral hip stem used in hip replacement surgery. The relatively rough surface of an implant is a favorable environment for bacteria to attach and grow. This can lead to the development of biofilms, which can cause infections. The researchers who took this image are working to understand where biofilms are likely to develop. This knowledge could support the prevention and treatment of infections. A scanning electron microscope was used to capture this image.
More information on the research that produced this image can be found in the Antibiotics paper "Free-floating aggregate and single-cell-initiated biofilms of Staphylococcus aureus" by Gupta et al.
Related to image 6803 and video 6805.
More information on the research that produced this image can be found in the Antibiotics paper "Free-floating aggregate and single-cell-initiated biofilms of Staphylococcus aureus" by Gupta et al.
Related to image 6803 and video 6805.
Paul Stoodley, The Ohio State University.
View Media
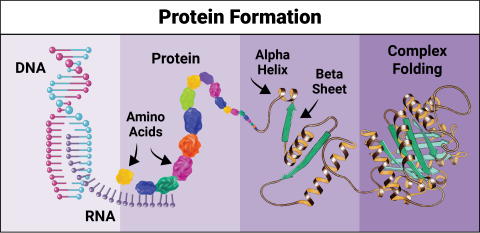
6603: Protein formation
6603: Protein formation
Proteins are 3D structures made up of smaller units. DNA is transcribed to RNA, which in turn is translated into amino acids. Amino acids form a protein strand, which has sections of corkscrew-like coils, called alpha helices, and other sections that fold flat, called beta sheets. The protein then goes through complex folding to produce the 3D structure.
NIGMS, with the folded protein illustration adapted from Jane Richardson, Duke University Medical Center
View Media
