Switch to List View
Image and Video Gallery
This is a searchable collection of scientific photos, illustrations, and videos. The images and videos in this gallery are licensed under Creative Commons Attribution Non-Commercial ShareAlike 3.0. This license lets you remix, tweak, and build upon this work non-commercially, as long as you credit and license your new creations under identical terms.

3772: The Proteasome: The Cell's Trash Processor in Action
3772: The Proteasome: The Cell's Trash Processor in Action
Our cells are constantly removing and recycling molecular waste. This video shows one way cells process their trash.
View Media
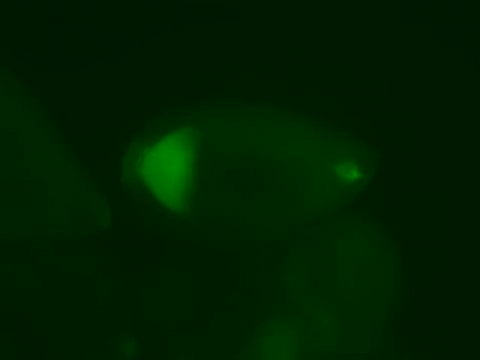
2315: Fly cells live
2315: Fly cells live
If a picture is worth a thousand words, what's a movie worth? For researchers studying cell migration, a "documentary" of fruit fly cells (bright green) traversing an egg chamber could answer longstanding questions about cell movement. Historically, researchers have been unable to watch this cell migration unfold in living ovarian tissue in real time. But by developing a culture medium that allows fly eggs to survive outside their ovarian homes, scientists can observe the nuances of cell migration as it happens. Such details may shed light on how immune cells move to a wound and why cancer cells spread to other sites. See 3594 for still image.
Denise Montell, Johns Hopkins University School of Medicine
View Media
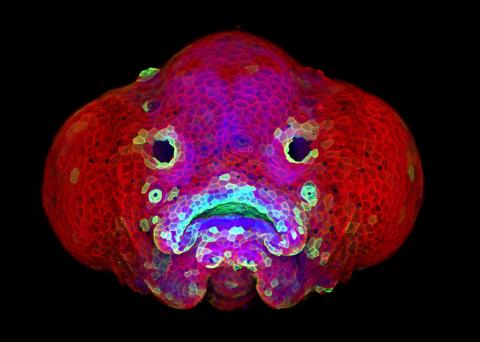
5881: Zebrafish larva
5881: Zebrafish larva
You are face to face with a 6-day-old zebrafish larva. What look like eyes will become nostrils, and the bulges on either side will become eyes. Scientists use fast-growing, transparent zebrafish to see body shapes form and organs develop over the course of just a few days. Images like this one help researchers understand how gene mutations can lead to facial abnormalities such as cleft lip and palate in people.
This image won a 2016 FASEB BioArt award. In addition, NIH Director Francis Collins featured this on his blog on January 26, 2017.
This image won a 2016 FASEB BioArt award. In addition, NIH Director Francis Collins featured this on his blog on January 26, 2017.
Oscar Ruiz and George Eisenhoffer, University of Texas MD Anderson Cancer Center, Houston
View Media
6489: CRISPR Illustration Frame 5
6489: CRISPR Illustration Frame 5
This illustration shows, in simplified terms, how the CRISPR-Cas9 system can be used as a gene-editing tool. This is the fifthframe in a series of five. The CRISPR system has two components joined together: a finely tuned targeting device (a small strand of RNA programmed to look for a specific DNA sequence) and a strong cutting device (an enzyme called Cas9 that can cut through a double strand of DNA). For an explanation and overview of the CRISPR-Cas9 system, see the NIGMS Biomedical Beat blog entry, Field Focus: Precision Gene Editing with CRISPR and the iBiology video, Genome Engineering with CRISPR-Cas9: Birth of a Breakthrough Technology.
View Media

5753: Clathrin-mediated endocytosis
5753: Clathrin-mediated endocytosis
Endocytosis is the process by which cells are able to take up membrane and extracellular materials through the formation of a small intracellular bubble, called a vesicle. This process, called membrane budding, is generally by a coating of proteins. This protein coat helps both to deform the membrane and to concentrate specific proteins inside the newly forming vesicle. Clathrin is a coat protein that functions in receptor-mediated endocytosis events at the plasma membrane. This animation shows the process of clathrin-mediated endocytosis. An iron-transport protein called transferrin (blue) is bound to its receptor (purple) on the exterior cell membrane. Inside the cell, a clathrin cage (shown in white/beige) assembles through interactions with membrane-bound adaptor proteins (green), causing the cell membrane to begin bending. The adaptor proteins also bind to receptors for transferrin, capturing them in the growing vesicle. Molecules of a protein called dynamin (purple) are then recruited to the neck of the vesicle and are involved in separating the membranes of the cell and the vesicle. Soon after the vesicle has budded off the membrane, the clathrin cage is disassembled. This disassembly is mediated by another protein called HSC70 (yellow), and its cofactor protein auxilin (orange).
Janet Iwasa, University of Utah
View Media
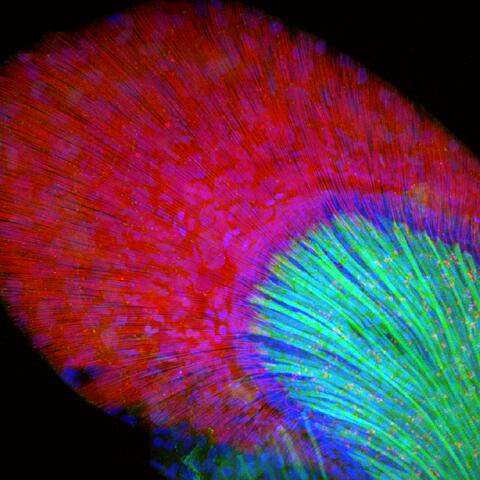
3598: Developing zebrafish fin
3598: Developing zebrafish fin
Originally from the waters of India, Nepal, and neighboring countries, zebrafish can now be found swimming in science labs (and home aquariums) throughout the world. This fish is a favorite study subject for scientists interested in how genes guide the early stages of prenatal development (including the developing fin shown here) and in the effects of environmental contamination on embryos.
In this image, green fluorescent protein (GFP) is expressed where the gene sox9b is expressed. Collagen (red) marks the fin rays, and DNA, stained with a dye called DAPI, is in blue. sox9b plays many important roles during development, including the building of the heart and brain, and is also necessary for skeletal development. At the University of Wisconsin, researchers have found that exposure to contaminants that bind the aryl-hydrocarbon receptor results in the downregulation of sox9b. Loss of sox9b severely disrupts development in zebrafish and causes a life-threatening disorder called campomelic dysplasia (CD) in humans. CD is characterized by cardiovascular, neural, and skeletal defects. By studying the roles of genes such as sox9b in zebrafish, scientists hope to better understand normal development in humans as well as how to treat developmental disorders and diseases.
This image was part of the Life: Magnified exhibit that ran from June 3, 2014, to January 21, 2015, at Dulles International Airport.
In this image, green fluorescent protein (GFP) is expressed where the gene sox9b is expressed. Collagen (red) marks the fin rays, and DNA, stained with a dye called DAPI, is in blue. sox9b plays many important roles during development, including the building of the heart and brain, and is also necessary for skeletal development. At the University of Wisconsin, researchers have found that exposure to contaminants that bind the aryl-hydrocarbon receptor results in the downregulation of sox9b. Loss of sox9b severely disrupts development in zebrafish and causes a life-threatening disorder called campomelic dysplasia (CD) in humans. CD is characterized by cardiovascular, neural, and skeletal defects. By studying the roles of genes such as sox9b in zebrafish, scientists hope to better understand normal development in humans as well as how to treat developmental disorders and diseases.
This image was part of the Life: Magnified exhibit that ran from June 3, 2014, to January 21, 2015, at Dulles International Airport.
Jessica Plavicki
View Media
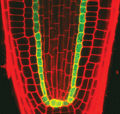
2329: Planting roots
2329: Planting roots
At the root tips of the mustard plant Arabidopsis thaliana (red), two proteins work together to control the uptake of water and nutrients. When the cell division-promoting protein called Short-root moves from the center of the tip outward, it triggers the production of another protein (green) that confines Short-root to the nutrient-filtering endodermis. The mechanism sheds light on how genes and proteins interact in a model organism and also could inform the engineering of plants.
Philip Benfey, Duke University
View Media
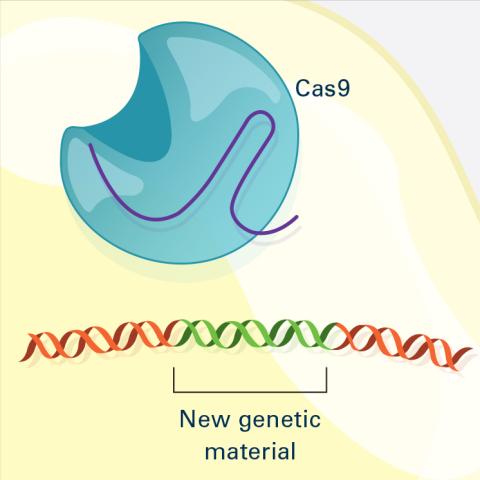
6488: CRISPR Illustration Frame 4
6488: CRISPR Illustration Frame 4
This illustration shows, in simplified terms, how the CRISPR-Cas9 system can be used as a gene-editing tool. The CRISPR system has two components joined together: a finely tuned targeting device (a small strand of RNA programmed to look for a specific DNA sequence) and a strong cutting device (an enzyme called Cas9 that can cut through a double strand of DNA). This frame (4 out of 4) shows a repaired DNA strand with new genetic material that researchers can introduce, which the cell automatically incorporates into the gap when it repairs the broken DNA.
For an explanation and overview of the CRISPR-Cas9 system, see the iBiology video, and find the full CRIPSR illustration here.
For an explanation and overview of the CRISPR-Cas9 system, see the iBiology video, and find the full CRIPSR illustration here.
National Institute of General Medical Sciences.
View Media

6356: H1N1 Influenza Virus
6356: H1N1 Influenza Virus
Related to image 6355.
Dr. Rommie Amaro, University of California, San Diego
View Media
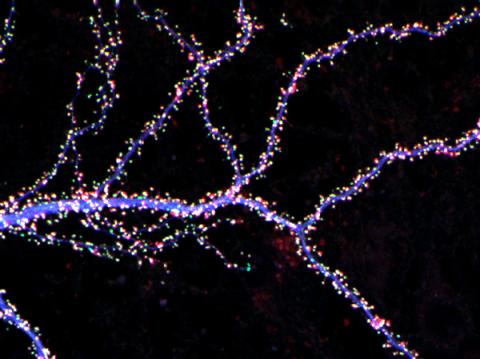
3686: Hippocampal neuron from rodent brain
3686: Hippocampal neuron from rodent brain
Hippocampal neuron from rodent brain with dendrites shown in blue. The hundreds of tiny magenta, green and white dots are the dendritic spines of excitatory synapses.
Shelley Halpain, UC San Diego
View Media
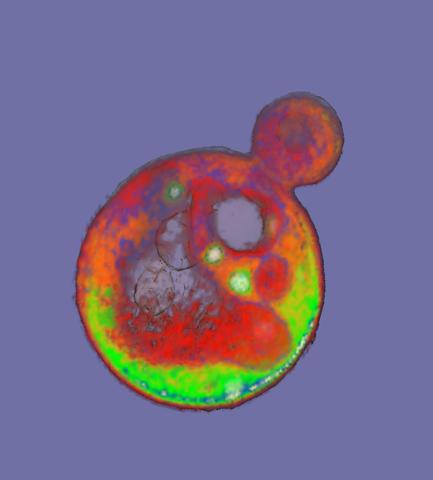
2307: Cells frozen in time
2307: Cells frozen in time
The fledgling field of X-ray microscopy lets researchers look inside whole cells rapidly frozen to capture their actions at that very moment. Here, a yeast cell buds before dividing into two. Colors show different parts of the cell. Seeing whole cells frozen in time will help scientists observe cells' complex structures and follow how molecules move inside them.
Carolyn Larabell, University of California, San Francisco, and the Lawrence Berkeley National Laboratory
View Media
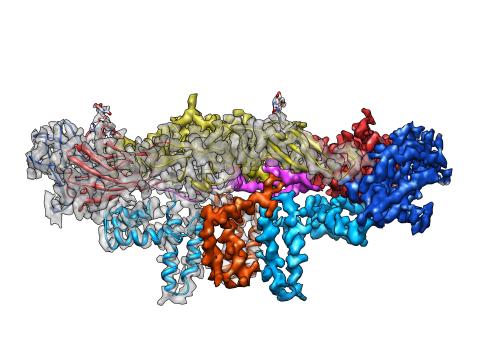
3758: Dengue virus membrane protein structure
3758: Dengue virus membrane protein structure
Dengue virus is a mosquito-borne illness that infects millions of people in the tropics and subtropics each year. Like many viruses, dengue is enclosed by a protective membrane. The proteins that span this membrane play an important role in the life cycle of the virus. Scientists used cryo-EM to determine the structure of a dengue virus at a 3.5-angstrom resolution to reveal how the membrane proteins undergo major structural changes as the virus matures and infects a host. The image shows a side view of the structure of a protein composed of two smaller proteins, called E and M. Each E and M contributes two molecules to the overall protein structure (called a heterotetramer), which is important for assembling and holding together the viral membrane, i.e., the shell that surrounds the genetic material of the dengue virus. The dengue protein's structure has revealed some portions in the protein that might be good targets for developing medications that could be used to combat dengue virus infections. For more on cryo-EM see the blog post Cryo-Electron Microscopy Reveals Molecules in Ever Greater Detail. You can watch a rotating view of the dengue virus surface structure in video 3748.
Hong Zhou, UCLA
View Media

2411: Fungal lipase (2)
2411: Fungal lipase (2)
Crystals of fungal lipase protein created for X-ray crystallography, which can reveal detailed, three-dimensional protein structures.
Alex McPherson, University of California, Irvine
View Media
6487: CRISPR Illustration Frame 3
6487: CRISPR Illustration Frame 3
This illustration shows, in simplified terms, how the CRISPR-Cas9 system can be used as a gene-editing tool. The CRISPR system has two components joined together: a finely tuned targeting device (a small strand of RNA programmed to look for a specific DNA sequence) and a strong cutting device (an enzyme called Cas9 that can cut through a double strand of DNA). In this frame (3 of 4), the Cas9 enzyme cuts both strands of the DNA.
For an explanation and overview of the CRISPR-Cas9 system, see the iBiology video, and find the full CRIPSR illustration here.
For an explanation and overview of the CRISPR-Cas9 system, see the iBiology video, and find the full CRIPSR illustration here.
National Institute of General Medical Sciences.
View Media
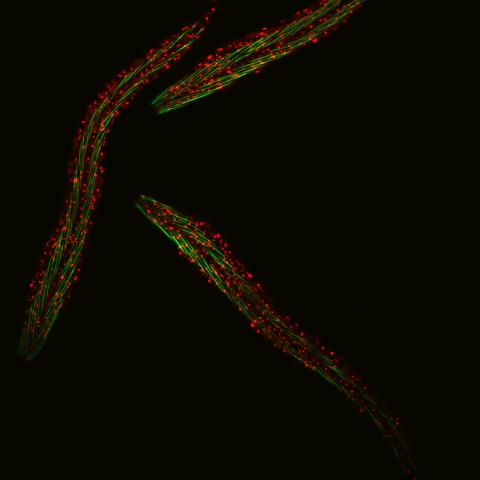
6582: Group of fluorescent C. elegans showing muscle and ribosomal protein
6582: Group of fluorescent C. elegans showing muscle and ribosomal protein
Three C. elegans, tiny roundworms, with a ribosomal protein glowing red and muscle fibers glowing green. Researchers used these worms to study a molecular pathway that affects aging. The ribosomal protein is involved in protein translation and may play a role in dietary restriction-induced longevity. Image created using confocal microscopy.
View single roundworm here 6581.
View closeup of roundworms here 6583.
View single roundworm here 6581.
View closeup of roundworms here 6583.
Jarod Rollins, Mount Desert Island Biological Laboratory.
View Media

2723: iPS cell facility at the Coriell Institute for Medical Research
2723: iPS cell facility at the Coriell Institute for Medical Research
This lab space was designed for work on the induced pluripotent stem (iPS) cell collection, part of the NIGMS Human Genetic Cell Repository at the Coriell Institute for Medical Research.
Courtney Sill, Coriell Institute for Medical Research
View Media
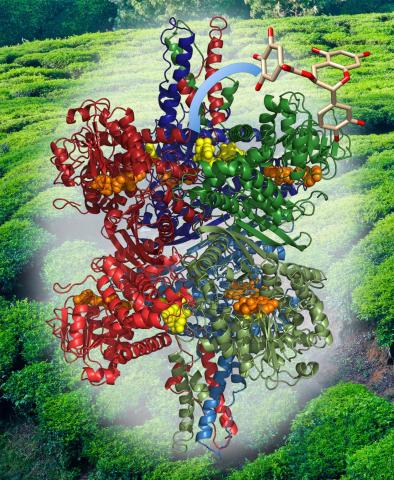
3421: Structure of Glutamate Dehydrogenase
3421: Structure of Glutamate Dehydrogenase
Some children are born with a mutation in a regulatory site on this enzyme that causes them to over-secrete insulin when they consume protein. We found that a compound from green tea (shown in the stick figure and by the yellow spheres on the enzyme) is able to block this hyperactivity when given to animals with this disorder.
Judy Coyle, Donald Danforth Plant Science Center
View Media

3344: Artificial cilia exhibit spontaneous beating
3344: Artificial cilia exhibit spontaneous beating
Researchers have created artificial cilia that wave like the real thing. Zvonimir Dogic and his Brandeis University colleagues combined just a few cilia proteins to create cilia that are able to wave and sweep material around--although more slowly and simply than real ones. The researchers are using the lab-made cilia to study how the structures coordinate their movements and what happens when they don't move properly. Featured in the August 18, 2011, issue of Biomedical Beat.
Zvonimir Dogic
View Media
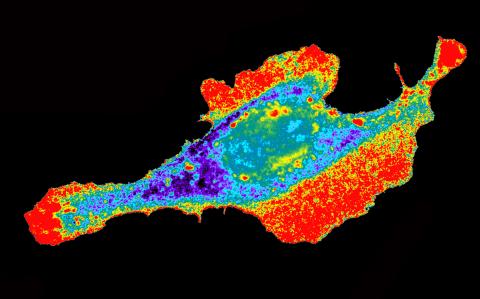
2453: Seeing signaling protein activation in cells 03
2453: Seeing signaling protein activation in cells 03
Cdc42, a member of the Rho family of small guanosine triphosphatase (GTPase) proteins, regulates multiple cell functions, including motility, proliferation, apoptosis, and cell morphology. In order to fulfill these diverse roles, the timing and location of Cdc42 activation must be tightly controlled. Klaus Hahn and his research group use special dyes designed to report protein conformational changes and interactions, here in living neutrophil cells. Warmer colors in this image indicate higher levels of activation. Cdc42 looks to be activated at cell protrusions.
Related to images 2451, 2452, and 2454.
Related to images 2451, 2452, and 2454.
Klaus Hahn, University of North Carolina, Chapel Hill Medical School
View Media

7011: Hawaiian bobtail squid
7011: Hawaiian bobtail squid
An adult Hawaiian bobtail squid, Euprymna scolopes, swimming next to a submerged hand.
Related to image 7010 and video 7012.
Related to image 7010 and video 7012.
Margaret J. McFall-Ngai, Carnegie Institution for Science/California Institute of Technology, and Edward G. Ruby, California Institute of Technology.
View Media
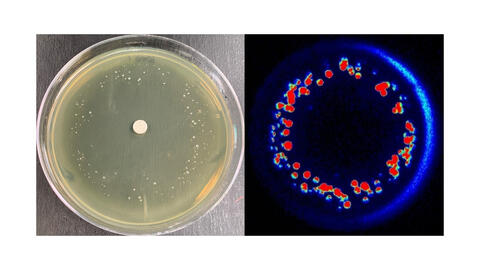
6802: Antibiotic-surviving bacteria
6802: Antibiotic-surviving bacteria
Colonies of bacteria growing despite high concentrations of antibiotics. These colonies are visible both by eye, as seen on the left, and by bioluminescence imaging, as seen on the right. The bioluminescent color indicates the metabolic activity of these bacteria, with their red centers indicating high metabolism.
More information about the research that produced this image can be found in the Antimicrobial Agents and Chemotherapy paper “Novel aminoglycoside-tolerant phoenix colony variants of Pseudomonas aeruginosa” by Sindeldecker et al.
More information about the research that produced this image can be found in the Antimicrobial Agents and Chemotherapy paper “Novel aminoglycoside-tolerant phoenix colony variants of Pseudomonas aeruginosa” by Sindeldecker et al.
Paul Stoodley, The Ohio State University.
View Media
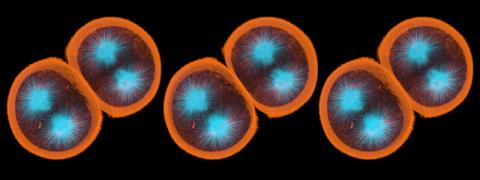
1047: Sea urchin embryo 01
1047: Sea urchin embryo 01
Stereo triplet of a sea urchin embryo stained to reveal actin filaments (orange) and microtubules (blue). This image is part of a series of images: image 1048, image 1049, image 1050, image 1051 and image 1052.
George von Dassow, University of Washington
View Media
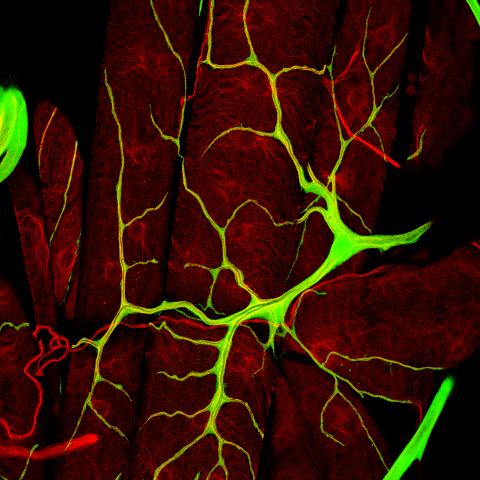
3615: An insect tracheal cell delivers air to muscles
3615: An insect tracheal cell delivers air to muscles
Insects like the fruit fly use an elaborate network of branching tubes called trachea (green) to transport oxygen throughout their bodies. Fruit flies have been used in biomedical research for more than 100 years and remain one of the most frequently studied model organisms. They have a large percentage of genes in common with us, including hundreds of genes that are associated with human diseases.
This image was part of the Life: Magnified exhibit that ran from June 3, 2014, to January 21, 2015, at Dulles International Airport.
This image was part of the Life: Magnified exhibit that ran from June 3, 2014, to January 21, 2015, at Dulles International Airport.
Jayan Nair and Maria Leptin, European Molecular Biology Laboratory, Heidelberg, Germany
View Media

2376: Protein purification facility
2376: Protein purification facility
The Center for Eukaryotic Structural Genomics protein purification facility is responsible for purifying all recombinant proteins produced by the center. The facility performs several purification steps, monitors the quality of the processes, and stores information about the biochemical properties of the purified proteins in the facility database.
Center for Eukaryotic Structural Genomics
View Media
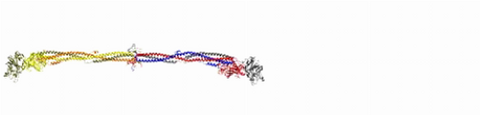
2450: Blood clots show their flex
2450: Blood clots show their flex
Blood clots stop bleeding, but they also can cause heart attacks and strokes. A team led by computational biophysicist Klaus Schulten of the University of Illinois at Urbana-Champaign has revealed how a blood protein can give clots their lifesaving and life-threatening abilities. The researchers combined experimental and computational methods to animate fibrinogen, a protein that forms the elastic fibers that enable clots to withstand the force of blood pressure. This simulation shows that the protein, through a series of events, stretches up to three times its length. Adjusting this elasticity could improve how we manage healthful and harmful clots. NIH's National Center for Research Resources also supported this work. Featured in the March 19, 2008, issue of Biomedical Beat.
Eric Lee, University of Illinois at Urbana-Champaign
View Media
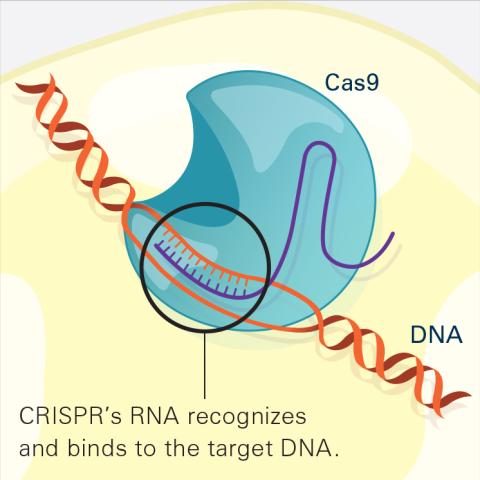
6486: CRISPR Illustration Frame 2
6486: CRISPR Illustration Frame 2
This illustration shows, in simplified terms, how the CRISPR-Cas9 system can be used as a gene-editing tool. The CRISPR system has two components joined together: a finely tuned targeting device (a small strand of RNA programmed to look for a specific DNA sequence) and a strong cutting device (an enzyme called Cas9 that can cut through a double strand of DNA). In this frame (2 of 4), the CRISPR machine locates the target DNA sequence once inserted into a cell.
For an explanation and overview of the CRISPR-Cas9 system, see the iBiology video, and find the full CRIPSR illustration here.
For an explanation and overview of the CRISPR-Cas9 system, see the iBiology video, and find the full CRIPSR illustration here.
National Institute of General Medical Sciences.
View Media
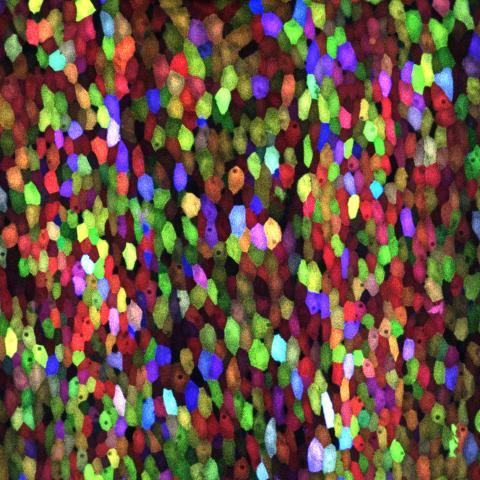
3782: A multicolored fish scale 1
3782: A multicolored fish scale 1
Each of the colored specs in this image is a cell on the surface of a fish scale. To better understand how wounds heal, scientists have inserted genes that make cells brightly glow in different colors into the skin cells of zebrafish, a fish often used in laboratory research. The colors enable the researchers to track each individual cell, for example, as it moves to the location of a cut or scrape over the course of several days. These technicolor fish endowed with glowing skin cells dubbed "skinbow" provide important insight into how tissues recover and regenerate after an injury.
For more information on skinbow fish, see the Biomedical Beat blog post Visualizing Skin Regeneration in Real Time and a press release from Duke University highlighting this research. Related to image 3783.
For more information on skinbow fish, see the Biomedical Beat blog post Visualizing Skin Regeneration in Real Time and a press release from Duke University highlighting this research. Related to image 3783.
Chen-Hui Chen and Kenneth Poss, Duke University
View Media
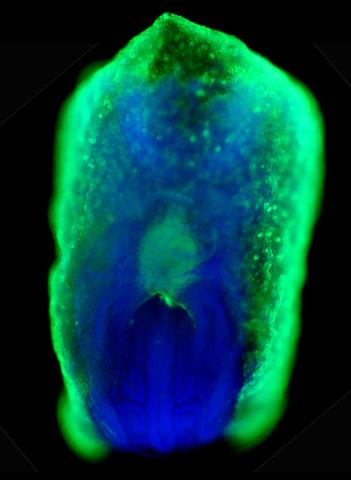
2607: Mouse embryo showing Smad4 protein
2607: Mouse embryo showing Smad4 protein
This eerily glowing blob isn't an alien or a creature from the deep sea--it's a mouse embryo just eight and a half days old. The green shell and core show a protein called Smad4. In the center, Smad4 is telling certain cells to begin forming the mouse's liver and pancreas. Researchers identified a trio of signaling pathways that help switch on Smad4-making genes, starting immature cells on the path to becoming organs. The research could help biologists learn how to grow human liver and pancreas tissue for research, drug testing and regenerative medicine. In addition to NIGMS, NIH's National Institute of Diabetes and Digestive and Kidney Diseases also supported this work.
Kenneth Zaret, Fox Chase Cancer Center
View Media
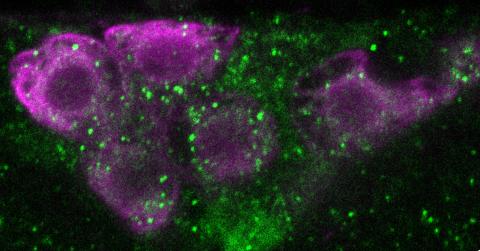
6982: Insulin production and fat sensing in fruit flies
6982: Insulin production and fat sensing in fruit flies
Fourteen neurons (magenta) in the adult Drosophila brain produce insulin, and fat tissue sends packets of lipids to the brain via the lipoprotein carriers (green). This image was captured using a confocal microscope and shows a maximum intensity projection of many slices.
Related to images 6983, 6984, and 6985.
Related to images 6983, 6984, and 6985.
Akhila Rajan, Fred Hutchinson Cancer Center
View Media
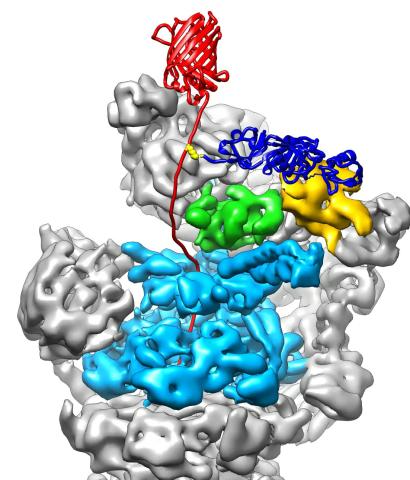
3763: The 26S proteasome engages with a protein substrate
3763: The 26S proteasome engages with a protein substrate
The proteasome is a critical multiprotein complex in the cell that breaks down and recycles proteins that have become damaged or are no longer needed. This illustration shows a protein substrate (red) that is bound through its ubiquitin chain (blue) to one of the ubiquitin receptors of the proteasome (Rpn10, yellow). The substrate's flexible engagement region gets engaged by the AAA+ motor of the proteasome (cyan), which initiates mechanical pulling, unfolding and movement of the protein into the proteasome's interior for cleavage into small shorter protein pieces called peptides. During movement of the substrate, its ubiquitin modification gets cleaved off by the deubiquitinase Rpn11 (green), which sits directly above the entrance to the AAA+ motor pore and acts as a gatekeeper to ensure efficient ubiquitin removal, a prerequisite for fast protein breakdown by the 26S proteasome. Related to video 3764.
Andreas Martin, HHMI
View Media
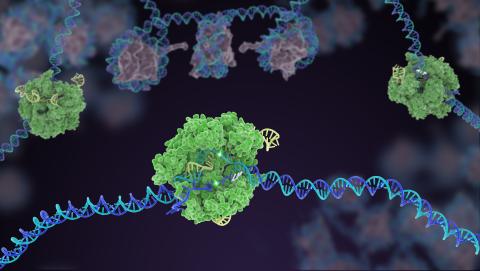
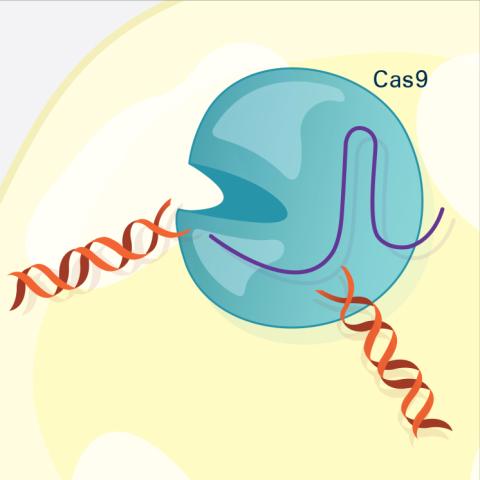
5816: Cas9 protein involved in the CRISPR gene-editing technology
5816: Cas9 protein involved in the CRISPR gene-editing technology
In the gene-editing tool CRISPR, a small strand of RNA identifies a specific chunk of DNA. Then the enzyme Cas9 (green) swoops in and cuts the double-stranded DNA (blue/purple) in two places, removing the specific chunk.
Janet Iwasa
View Media

3735: Scanning electron microscopy of collagen fibers
3735: Scanning electron microscopy of collagen fibers
This image shows collagen, a fibrous protein that's the main component of the extracellular matrix (ECM). Collagen is a strong, ropelike molecule that forms stretch-resistant fibers. The most abundant protein in our bodies, collagen accounts for about a quarter of our total protein mass. Among its many functions is giving strength to our tendons, ligaments and bones and providing scaffolding for skin wounds to heal. There are about 20 different types of collagen in our bodies, each adapted to the needs of specific tissues.
Tom Deerinck, National Center for Microscopy and Imaging Research (NCMIR)
View Media
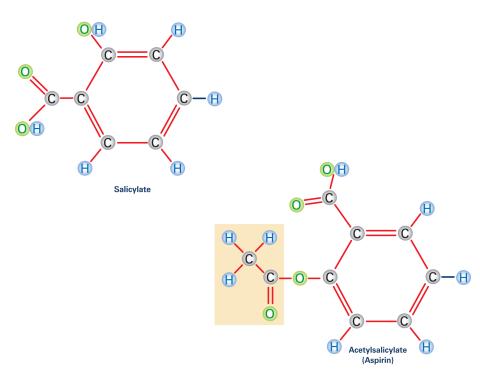
2530: Aspirin (with labels)
2530: Aspirin (with labels)
Acetylsalicylate (bottom) is the aspirin of today. Adding a chemical tag called an acetyl group (shaded box, bottom) to a molecule derived from willow bark (salicylate, top) makes the molecule less acidic (and easier on the lining of the digestive tract), but still effective at relieving pain. See image 2529 for an unlabeled version of this illustration. Featured in Medicines By Design.
Crabtree + Company
View Media
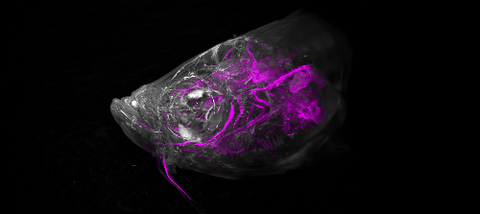
6933: Zebrafish head vasculature video
6933: Zebrafish head vasculature video
Various views of a zebrafish head with blood vessels shown in purple. Researchers often study zebrafish because they share many genes with humans, grow and reproduce quickly, and have see-through eggs and embryos, which make it easy to study early stages of development.
This video was captured using a light sheet microscope.
Related to image 6934.
This video was captured using a light sheet microscope.
Related to image 6934.
Prayag Murawala, MDI Biological Laboratory and Hannover Medical School.
View Media
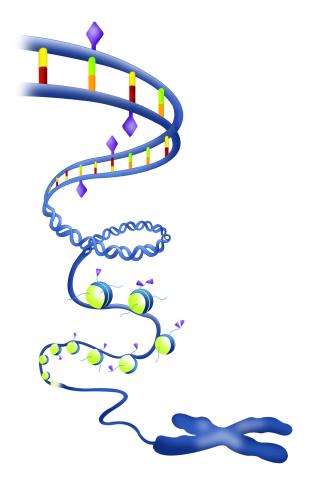
2562: Epigenetic code
2562: Epigenetic code
The "epigenetic code" controls gene activity with chemical tags that mark DNA (purple diamonds) and the "tails" of histone proteins (purple triangles). These markings help determine whether genes will be transcribed by RNA polymerase. Genes hidden from access to RNA polymerase are not expressed. See image 2563 for a labeled version of this illustration. Featured in The New Genetics.
Crabtree + Company
View Media
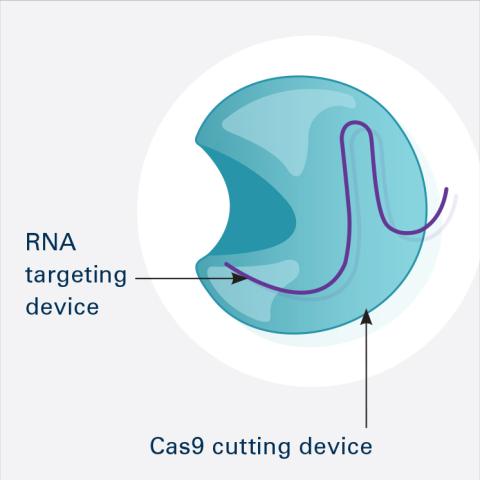
6465: CRISPR Illustration Frame 1
6465: CRISPR Illustration Frame 1
This illustration shows, in simplified terms, how the CRISPR-Cas9 system can be used as a gene-editing tool. This is the first frame in a series of four. The CRISPR system has two components joined together: a finely tuned targeting device (a small strand of RNA programmed to look for a specific DNA sequence) and a strong cutting device (an enzyme called Cas9 that can cut through a double strand of DNA).
For an explanation and overview of the CRISPR-Cas9 system, see the iBiology video, and find the full CRIPSR illustration here.
For an explanation and overview of the CRISPR-Cas9 system, see the iBiology video, and find the full CRIPSR illustration here.
National Institute of General Medical Sciences.
View Media
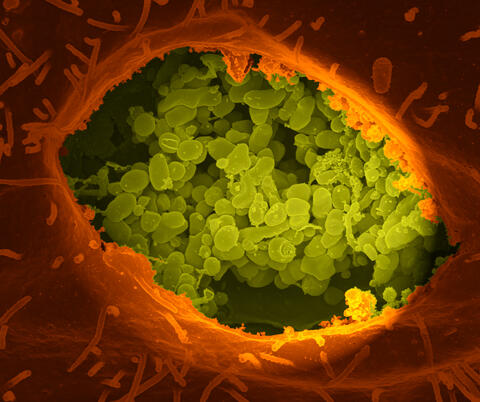
3621: Q fever bacteria in an infected cell
3621: Q fever bacteria in an infected cell
This image shows Q fever bacteria (yellow), which infect cows, sheep, and goats around the world and can infect humans, as well. When caught early, Q fever can be cured with antibiotics. A small fraction of people can develop a more serious, chronic form of the disease.
This image was part of the Life: Magnified exhibit that ran from June 3, 2014, to January 21, 2015, at Dulles International Airport.
This image was part of the Life: Magnified exhibit that ran from June 3, 2014, to January 21, 2015, at Dulles International Airport.
Robert Heinzen, Elizabeth Fischer, and Anita Mora, National Institute of Allergy and Infectious Diseases, National Institutes of Health
View Media
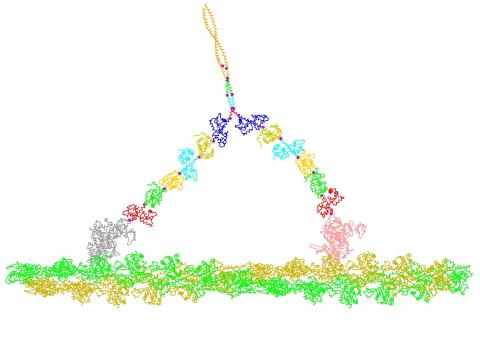
2754: Myosin V binding to actin
2754: Myosin V binding to actin
This simulation of myosin V binding to actin was created using the software tool Protein Mechanica. With Protein Mechanica, researchers can construct models using information from a variety of sources: crystallography, cryo-EM, secondary structure descriptions, as well as user-defined solid shapes, such as spheres and cylinders. The goal is to enable experimentalists to quickly and easily simulate how different parts of a molecule interact.
Simbios, NIH Center for Biomedical Computation at Stanford
View Media
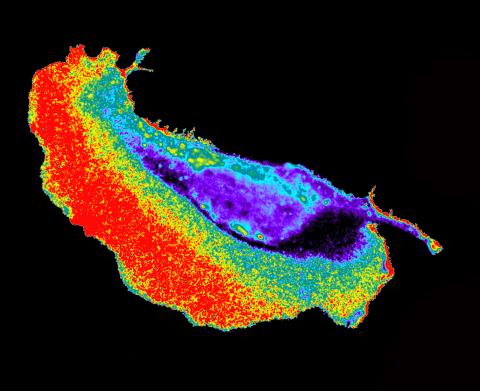
2452: Seeing signaling protein activation in cells 02
2452: Seeing signaling protein activation in cells 02
Cdc42, a member of the Rho family of small guanosine triphosphatase (GTPase) proteins, regulates multiple cell functions, including motility, proliferation, apoptosis, and cell morphology. In order to fulfill these diverse roles, the timing and location of Cdc42 activation must be tightly controlled. Klaus Hahn and his research group use special dyes designed to report protein conformational changes and interactions, here in living neutrophil cells. Warmer colors in this image indicate higher levels of activation. Cdc42 looks to be activated at cell protrusions.
Related to images 2451, 2453, and 2454.
Related to images 2451, 2453, and 2454.
Klaus Hahn, University of North Carolina, Chapel Hill Medical School
View Media
2442: Hydra 06
2442: Hydra 06
Hydra magnipapillata is an invertebrate animal used as a model organism to study developmental questions, for example the formation of the body axis.
Hiroshi Shimizu, National Institute of Genetics in Mishima, Japan
View Media
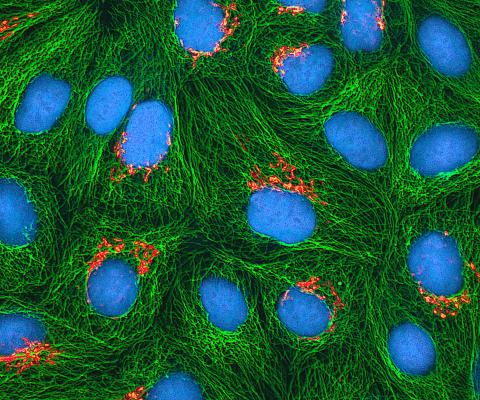
3522: HeLa cells
3522: HeLa cells
Multiphoton fluorescence image of cultured HeLa cells with a fluorescent protein targeted to the Golgi apparatus (orange), microtubules (green) and counterstained for DNA (cyan). Nikon RTS2000MP custom laser scanning microscope. See related images 3518, 3519, 3520, 3521.
National Center for Microscopy and Imaging Research (NCMIR)
View Media
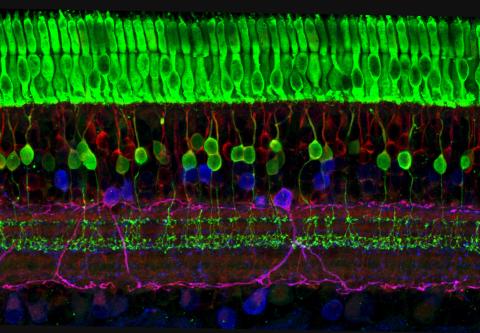
3635: The eye uses many layers of nerve cells to convert light into sight
3635: The eye uses many layers of nerve cells to convert light into sight
This image captures the many layers of nerve cells in the retina. The top layer (green) is made up of cells called photoreceptors that convert light into electrical signals to relay to the brain. The two best-known types of photoreceptor cells are rod- and cone-shaped. Rods help us see under low-light conditions but can't help us distinguish colors. Cones don't function well in the dark but allow us to see vibrant colors in daylight.
This image was part of the Life: Magnified exhibit that ran from June 3, 2014, to January 21, 2015, at Dulles International Airport.
This image was part of the Life: Magnified exhibit that ran from June 3, 2014, to January 21, 2015, at Dulles International Airport.
Wei Li, National Eye Institute, National Institutes of Health
View Media
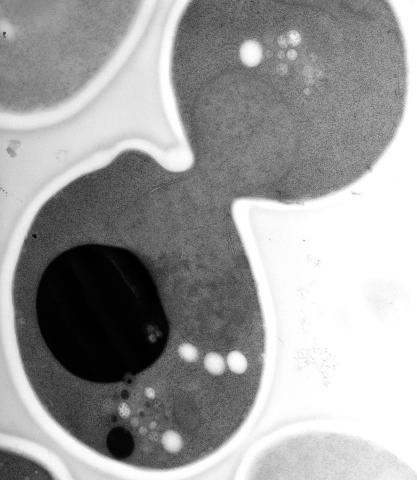
5770: EM of yeast cell division
5770: EM of yeast cell division
Cell division is an incredibly coordinated process. It not only ensures that the new cells formed during this event have a full set of chromosomes, but also that they are endowed with all the cellular materials, including proteins, lipids and small functional compartments called organelles, that are required for normal cell activity. This proper apportioning of essential cell ingredients helps each cell get off to a running start.
This image shows an electron microscopy (EM) thin section taken at 10,000x magnification of a dividing yeast cell over-expressing the protein ubiquitin, which is involved in protein degradation and recycling. The picture features mother and daughter endosome accumulations (small organelles with internal vesicles), a darkly stained vacuole and a dividing nucleus in close contact with a cadre of lipid droplets (unstained spherical bodies). Other dynamic events are also visible, such as spindle microtubules in the nucleus and endocytic pits at the plasma membrane.
These extensive details were revealed thanks to a preservation method involving high-pressure freezing, freeze-substitution and Lowicryl HM20 embedding.
This image shows an electron microscopy (EM) thin section taken at 10,000x magnification of a dividing yeast cell over-expressing the protein ubiquitin, which is involved in protein degradation and recycling. The picture features mother and daughter endosome accumulations (small organelles with internal vesicles), a darkly stained vacuole and a dividing nucleus in close contact with a cadre of lipid droplets (unstained spherical bodies). Other dynamic events are also visible, such as spindle microtubules in the nucleus and endocytic pits at the plasma membrane.
These extensive details were revealed thanks to a preservation method involving high-pressure freezing, freeze-substitution and Lowicryl HM20 embedding.
Matthew West and Greg Odorizzi, University of Colorado
View Media

6355: H1N1 Influenza Virus
6355: H1N1 Influenza Virus
CellPack image of the H1N1 influenza virus, with hemagglutinin and neuraminidase glycoproteins in green and red, respectively, on the outer envelope (white); matrix protein in gray, and ribonucleoprotein particles inside the virus in red and green. Related to image 6356.
Dr. Rommie Amaro, University of California, San Diego
View Media
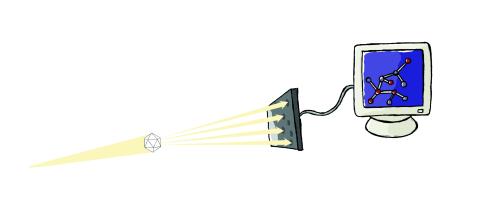
2511: X-ray crystallography
2511: X-ray crystallography
X-ray crystallography allows researchers to see structures too small to be seen by even the most powerful microscopes. To visualize the arrangement of atoms within molecules, researchers can use the diffraction patterns obtained by passing X-ray beams through crystals of the molecule. This is a common way for solving the structures of proteins. See image 2512 for a labeled version of this illustration. Featured in The Structures of Life.
Crabtree + Company
View Media

6887: Chromatin in human fibroblast
6887: Chromatin in human fibroblast
The nucleus of a human fibroblast cell with chromatin—a substance made up of DNA and proteins—shown in various colors. Fibroblasts are one of the most common types of cells in mammalian connective tissue, and they play a key role in wound healing and tissue repair. This image was captured using Stochastic Optical Reconstruction Microscopy (STORM).
Related to images 6888 and 6893.
Related to images 6888 and 6893.
Melike Lakadamyali, Perelman School of Medicine at the University of Pennsylvania.
View Media
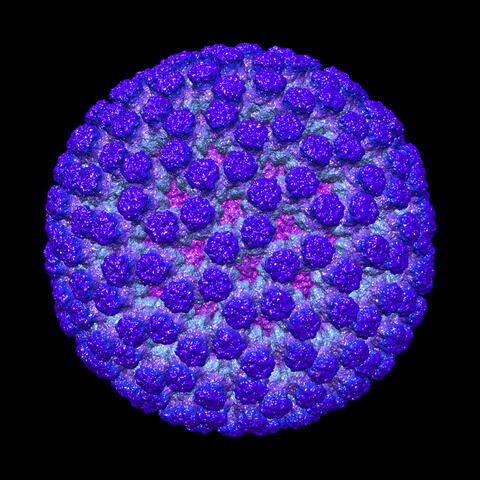
3584: Rotavirus structure
3584: Rotavirus structure
This image shows a computer-generated, three-dimensional map of the rotavirus structure. This virus infects humans and other animals and causes severe diarrhea in infants and young children. By the age of five, almost every child in the world has been infected with this virus at least once. Scientists have found a vaccine against rotavirus, so in the United States there are very few fatalities, but in developing countries and in places where the vaccine is unavailable, this virus is responsible for more than 200,000 deaths each year.
The rotavirus comprises three layers: the outer, middle and inner layers. On infection, the outer layer is removed, leaving behind a "double-layered particle." Researchers have studied the structure of this double-layered particle with a transmission electron microscope. Many images of the virus at a magnification of ~50,000x were acquired, and computational analysis was used to combine the individual particle images into a three-dimensional reconstruction.
The image was rendered by Melody Campbell (PhD student at TSRI). Work that led to the 3D map was published in Campbell et al. Movies of ice-embedded particles enhance resolution in electron cryo-microscopy. Structure. 2012;20(11):1823-8. PMCID: PMC3510009.
This image was part of the Life: Magnified exhibit that ran from June 3, 2014, to January 21, 2015, at Dulles International Airport.
The rotavirus comprises three layers: the outer, middle and inner layers. On infection, the outer layer is removed, leaving behind a "double-layered particle." Researchers have studied the structure of this double-layered particle with a transmission electron microscope. Many images of the virus at a magnification of ~50,000x were acquired, and computational analysis was used to combine the individual particle images into a three-dimensional reconstruction.
The image was rendered by Melody Campbell (PhD student at TSRI). Work that led to the 3D map was published in Campbell et al. Movies of ice-embedded particles enhance resolution in electron cryo-microscopy. Structure. 2012;20(11):1823-8. PMCID: PMC3510009.
This image was part of the Life: Magnified exhibit that ran from June 3, 2014, to January 21, 2015, at Dulles International Airport.
Bridget Carragher, The Scripps Research Institute, La Jolla, CA
View Media
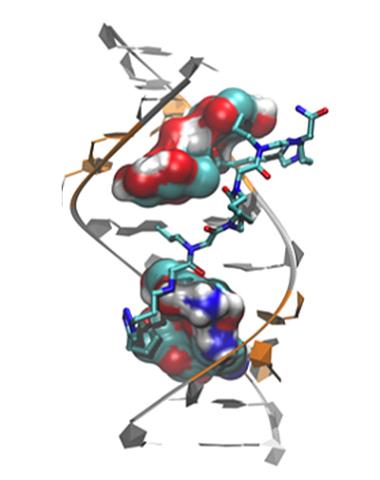
3573: Myotonic dystrophy type 2 genetic defect
3573: Myotonic dystrophy type 2 genetic defect
Scientists revealed a detailed image of the genetic defect that causes myotonic dystrophy type 2 and used that information to design drug candidates to counteract the disease.
Matthew Disney, Scripps Research Institute and Ilyas Yildirim, Northwestern University
View Media

6770: Group of Culex quinquefasciatus mosquito larvae
6770: Group of Culex quinquefasciatus mosquito larvae
Mosquito larvae with genes edited by CRISPR. This species of mosquito, Culex quinquefasciatus, can transmit West Nile virus, Japanese encephalitis virus, and avian malaria, among other diseases. The researchers who took this image developed a gene-editing toolkit for Culex quinquefasciatus that could ultimately help stop the mosquitoes from spreading pathogens. The work is described in the Nature Communications paper "Optimized CRISPR tools and site-directed transgenesis towards gene drive development in Culex quinquefasciatus mosquitoes" by Feng et al. Related to image 6769 and video 6771.
Valentino Gantz, University of California, San Diego.
View Media