Switch to List View
Image and Video Gallery
This is a searchable collection of scientific photos, illustrations, and videos. The images and videos in this gallery are licensed under Creative Commons Attribution Non-Commercial ShareAlike 3.0. This license lets you remix, tweak, and build upon this work non-commercially, as long as you credit and license your new creations under identical terms.
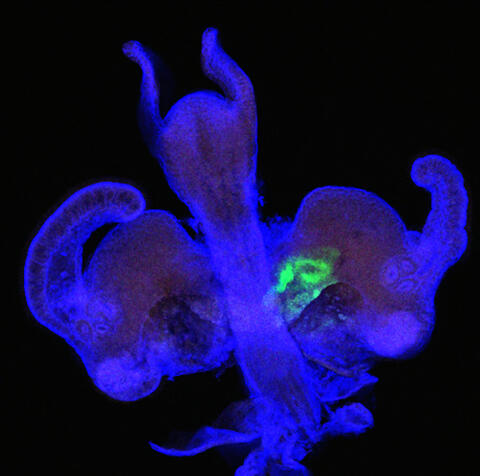
7020: Bacterial symbionts colonizing the crypts of a juvenile Hawaiian bobtail squid light organ
7020: Bacterial symbionts colonizing the crypts of a juvenile Hawaiian bobtail squid light organ
A light organ (~0.5 mm across) of a Hawaiian bobtail squid, Euprymna scolopes, stained blue. At the time of this image, the crypts within the tissues of only one side of the organ had been colonized by green-fluorescent protein-labeled Vibrio fischeri cells, which can be seen here in green. This image was taken using confocal fluorescence microscopy.
Related to images 7016, 7017, 7018, and 7019.
Related to images 7016, 7017, 7018, and 7019.
Margaret J. McFall-Ngai, Carnegie Institution for Science/California Institute of Technology, and Edward G. Ruby, California Institute of Technology.
View Media
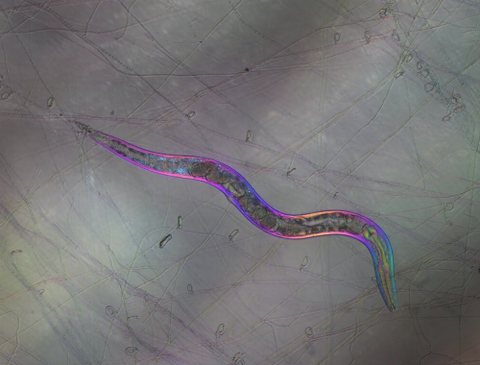
6963: C. elegans trapped by carnivorous fungus
6963: C. elegans trapped by carnivorous fungus
Real-time footage of Caenorhabditis elegans, a tiny roundworm, trapped by a carnivorous fungus, Arthrobotrys dactyloides. This fungus makes ring traps in response to the presence of C. elegans. When a worm enters a ring, the trap rapidly constricts so that the worm cannot move away, and the fungus then consumes the worm. The size of the imaged area is 0.7mm x 0.9mm.
This video was obtained with a polychromatic polarizing microscope (PPM) in white light that shows the polychromatic birefringent image with hue corresponding to the slow axis orientation. More information about PPM can be found in the Scientific Reports paper “Polychromatic Polarization Microscope: Bringing Colors to a Colorless World” by Shribak.
This video was obtained with a polychromatic polarizing microscope (PPM) in white light that shows the polychromatic birefringent image with hue corresponding to the slow axis orientation. More information about PPM can be found in the Scientific Reports paper “Polychromatic Polarization Microscope: Bringing Colors to a Colorless World” by Shribak.
Michael Shribak, Marine Biological Laboratory/University of Chicago.
View Media
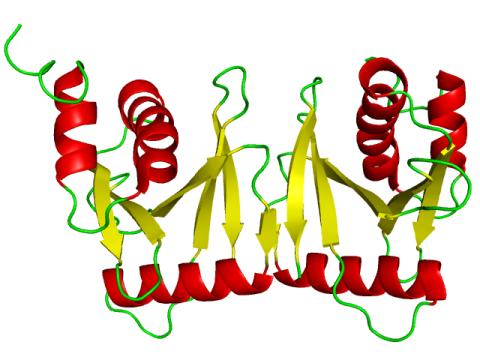
2351: tRNA splicing enzyme endonuclease in humans
2351: tRNA splicing enzyme endonuclease in humans
An NMR solution structure model of the transfer RNA splicing enzyme endonuclease in humans (subunit Sen15). This represents the first structure of a eukaryotic tRNA splicing endonuclease subunit.
Center for Eukaryotic Structural Genomics, PSI
View Media
3266: Biopixels
3266: Biopixels
Bioengineers were able to coax bacteria to blink in unison on microfluidic chips. This image shows a small chip with about 500 blinking bacterial colonies or biopixels. Related to images 3265 and 3268. From a UC San Diego news release, "Researchers create living 'neon signs' composed of millions of glowing bacteria."
Jeff Hasty Lab, UC San Diego
View Media
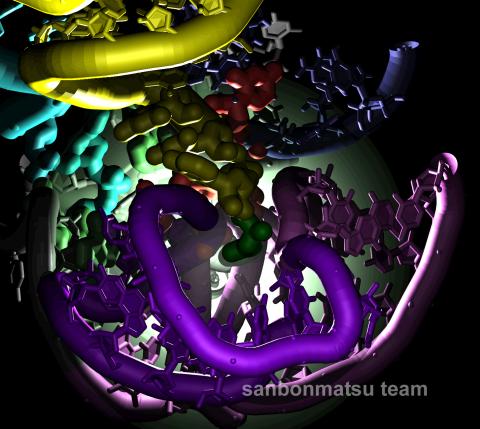
2336: Natural nanomachine in action
2336: Natural nanomachine in action
Using a supercomputer to simulate the movement of atoms in a ribosome, researchers looked into the core of this protein-making nanomachine and took snapshots. The picture shows an amino acid (green) being delivered by transfer RNA (yellow) into a corridor (purple) in the ribosome. In the corridor, a series of chemical reactions will string together amino acids to make a protein. The research project, which tracked the movement of more than 2.6 million atoms, was the largest computer simulation of a biological structure to date. The results shed light on the manufacturing of proteins and could aid the search for new antibiotics, which typically work by disabling the ribosomes of bacteria.
Kevin Sanbonmatsu, Los Alamos National Laboratory
View Media
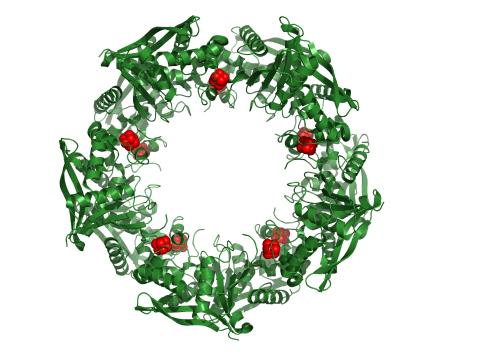
3720: Cas4 nuclease protein structure
3720: Cas4 nuclease protein structure
This wreath represents the molecular structure of a protein, Cas4, which is part of a system, known as CRISPR, that bacteria use to protect themselves against viral invaders. The green ribbons show the protein's structure, and the red balls show the location of iron and sulfur molecules important for the protein's function. Scientists harnessed Cas9, a different protein in the bacterial CRISPR system, to create a gene-editing tool known as CRISPR-Cas9. Using this tool, researchers are able to study a range of cellular processes and human diseases more easily, cheaply and precisely. In December, 2015, Science magazine recognized the CRISPR-Cas9 gene-editing tool as the "breakthrough of the year." Read more about Cas4 in the December 2015 Biomedical Beat post A Holiday-Themed Image Collection.
Fred Dyda, NIDDK
View Media
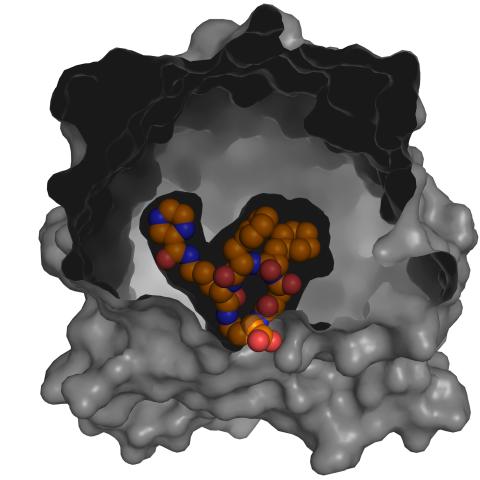
3417: X-ray co-crystal structure of Src kinase bound to a DNA-templated macrocycle inhibitor 5
3417: X-ray co-crystal structure of Src kinase bound to a DNA-templated macrocycle inhibitor 5
X-ray co-crystal structure of Src kinase bound to a DNA-templated macrocycle inhibitor. Related to images 3413, 3414, 3415, 3416, 3418, and 3419.
Markus A. Seeliger, Stony Brook University Medical School and David R. Liu, Harvard University
View Media
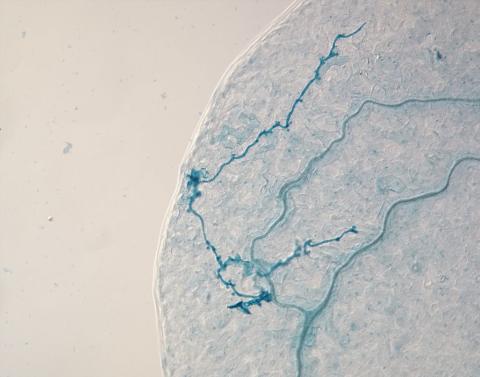
2780: Arabidopsis leaf injected with a pathogen
2780: Arabidopsis leaf injected with a pathogen
This is a magnified view of an Arabidopsis thaliana leaf eight days after being infected with the pathogen Hyaloperonospora arabidopsidis, which is closely related to crop pathogens that cause 'downy mildew' diseases. It is also more distantly related to the agent that caused the Irish potato famine. The veins of the leaf are light blue; in darker blue are the pathogen's hyphae growing through the leaf. The small round blobs along the length of the hyphae are called haustoria; each is invading a single plant cell to suck nutrients from the cell. Jeff Dangl and other NIGMS-supported researchers investigate how this pathogen and other like it use virulence mechanisms to suppress host defense and help the pathogens grow.
Jeff Dangl, University of North Carolina, Chapel Hill
View Media
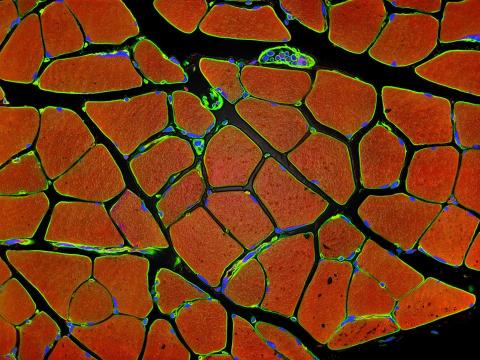
3677: Human skeletal muscle
3677: Human skeletal muscle
Cross section of human skeletal muscle. Image taken with a confocal fluorescent light microscope.
Tom Deerinck, National Center for Microscopy and Imaging Research (NCMIR)
View Media
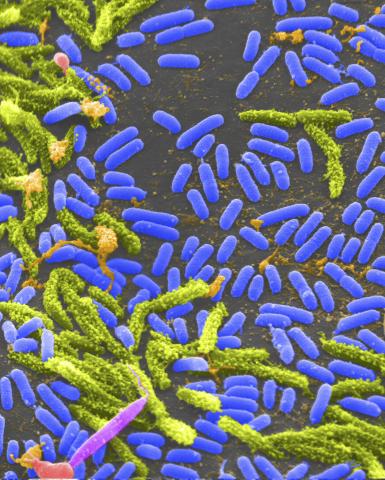
1160: Vibrio bacteria
1160: Vibrio bacteria
Vibrio, a type (genus) of rod-shaped bacteria. Some Vibrio species cause cholera in humans.
Tina Weatherby Carvalho, University of Hawaii at Manoa
View Media
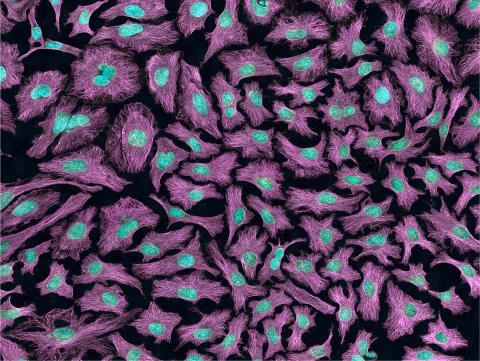
3520: HeLa cells
3520: HeLa cells
Multiphoton fluorescence image of HeLa cells with cytoskeletal microtubules (magenta) and DNA (cyan). Nikon RTS2000MP custom laser scanning microscope. See related images 3518, 3519, 3521, 3522.
National Center for Microscopy and Imaging Research (NCMIR)
View Media
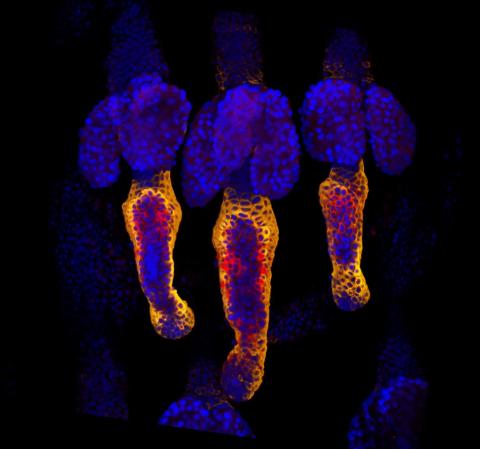
3499: Growing hair follicle stem cells
3499: Growing hair follicle stem cells
Wound healing requires the action of stem cells. In mice that lack the Sept2/ARTS gene, stem cells involved in wound healing live longer and wounds heal faster and more thoroughly than in normal mice. This confocal microscopy image from a mouse lacking the Sept2/ARTS gene shows a tail wound in the process of healing. Cell nuclei are in blue. Red and orange mark hair follicle stem cells (hair follicle stem cells activate to cause hair regrowth, which indicates healing). See more information in the article in Science.
Hermann Steller, Rockefeller University
View Media
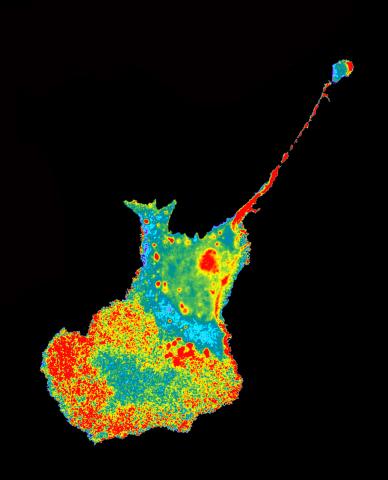
2454: Seeing signaling protein activation in cells 04
2454: Seeing signaling protein activation in cells 04
Cdc42, a member of the Rho family of small guanosine triphosphatase (GTPase) proteins, regulates multiple cell functions, including motility, proliferation, apoptosis, and cell morphology. In order to fulfill these diverse roles, the timing and location of Cdc42 activation must be tightly controlled. Klaus Hahn and his research group use special dyes designed to report protein conformational changes and interactions, here in living neutrophil cells. Warmer colors in this image indicate higher levels of activation. Cdc42 looks to be activated at cell protrusions.
Related to images 2451, 2452, and 2453.
Related to images 2451, 2452, and 2453.
Klaus Hahn, University of North Carolina, Chapel Hill Medical School
View Media
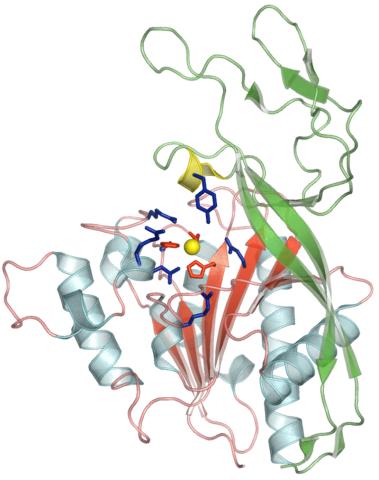
2352: Human aspartoacylase
2352: Human aspartoacylase
Model of aspartoacylase, a human enzyme involved in brain metabolism.
Center for Eukaryotic Structural Genomics, PSI
View Media
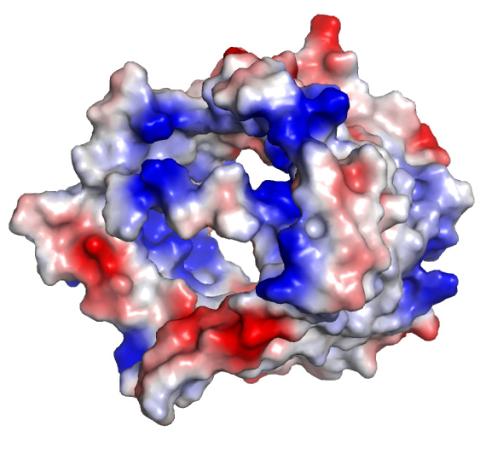
2494: VDAC-1 (3)
2494: VDAC-1 (3)
The structure of the pore-forming protein VDAC-1 from humans. This molecule mediates the flow of products needed for metabolism--in particular the export of ATP--across the outer membrane of mitochondria, the power plants for eukaryotic cells. VDAC-1 is involved in metabolism and the self-destruction of cells--two biological processes central to health.
Related to images 2491, 2495, and 2488.
Related to images 2491, 2495, and 2488.
Gerhard Wagner, Harvard Medical School
View Media
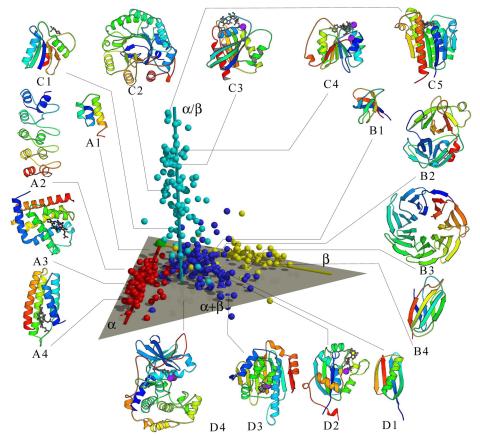
2367: Map of protein structures 02
2367: Map of protein structures 02
A global "map of the protein structure universe" indicating the positions of specific proteins. The preponderance of small, less-structured proteins near the origin, with the more highly structured, large proteins towards the ends of the axes, may suggest the evolution of protein structures.
Berkeley Structural Genomics Center, PSI
View Media
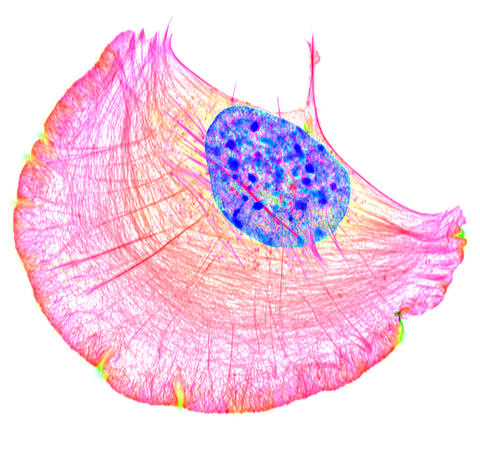
6964: Crawling cell
6964: Crawling cell
A crawling cell with DNA shown in blue and actin filaments, which are a major component of the cytoskeleton, visible in pink. Actin filaments help enable cells to crawl. This image was captured using structured illumination microscopy.
Dylan T. Burnette, Vanderbilt University School of Medicine.
View Media
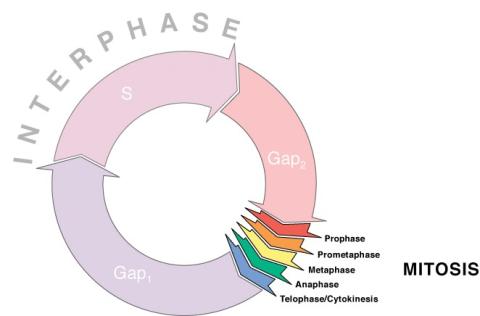
1310: Cell cycle wheel
1310: Cell cycle wheel
A typical animal cell cycle lasts roughly 24 hours, but depending on the type of cell, it can vary in length from less than 8 hours to more than a year. Most of the variability occurs in Gap1. Appears in the NIGMS booklet Inside the Cell.
Judith Stoffer
View Media
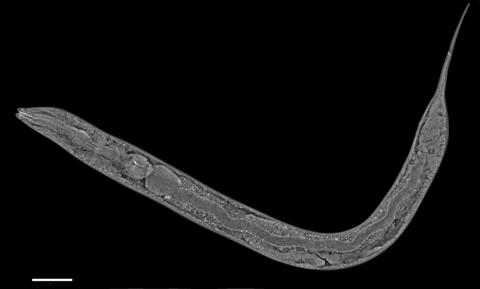
6961: C. elegans showing internal structures
6961: C. elegans showing internal structures
An image of Caenorhabditis elegans, a tiny roundworm, showing internal structures including the intestine, pharynx, and body wall muscle. C. elegans is one of the simplest organisms with a nervous system. Scientists use it to study nervous system development, among other things. This image was captured with a quantitative orientation-independent differential interference contrast (OI-DIC) microscope. The scale bar is 100 µm.
More information about the microscopy that produced this image can be found in the Journal of Microscopy paper by Malamy and Shribak.
More information about the microscopy that produced this image can be found in the Journal of Microscopy paper by Malamy and Shribak.
Michael Shribak, Marine Biological Laboratory/University of Chicago.
View Media
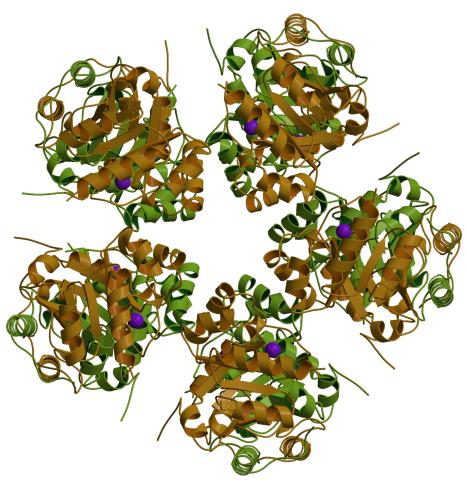
2380: PanB from M. tuberculosis (1)
2380: PanB from M. tuberculosis (1)
Model of an enzyme, PanB, from Mycobacterium tuberculosis, the bacterium that causes most cases of tuberculosis. This enzyme is an attractive drug target.
Mycobacterium Tuberculosis Center, PSI
View Media
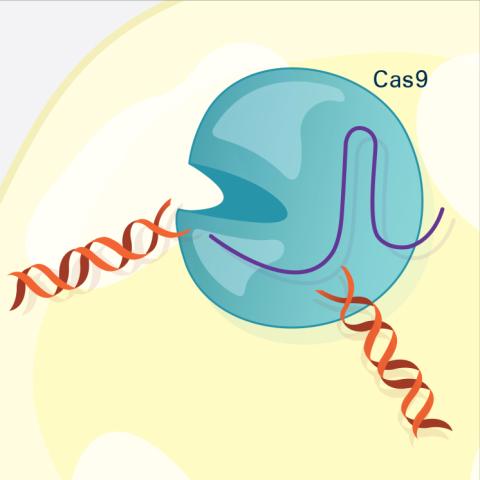
6487: CRISPR Illustration Frame 3
6487: CRISPR Illustration Frame 3
This illustration shows, in simplified terms, how the CRISPR-Cas9 system can be used as a gene-editing tool. The CRISPR system has two components joined together: a finely tuned targeting device (a small strand of RNA programmed to look for a specific DNA sequence) and a strong cutting device (an enzyme called Cas9 that can cut through a double strand of DNA). In this frame (3 of 4), the Cas9 enzyme cuts both strands of the DNA.
For an explanation and overview of the CRISPR-Cas9 system, see the iBiology video, and find the full CRIPSR illustration here.
For an explanation and overview of the CRISPR-Cas9 system, see the iBiology video, and find the full CRIPSR illustration here.
National Institute of General Medical Sciences.
View Media
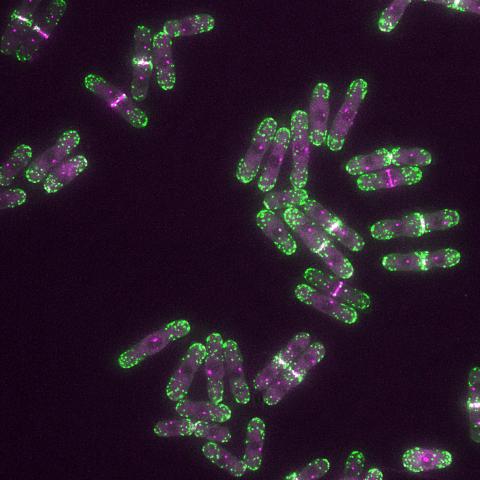
6793: Yeast cells with endocytic actin patches
6793: Yeast cells with endocytic actin patches
Yeast cells with endocytic actin patches (green). These patches help cells take in outside material. When a cell is in interphase, patches concentrate at its ends. During later stages of cell division, patches move to where the cell splits. This image was captured using wide-field microscopy with deconvolution.
Related to images 6791, 6792, 6794, 6797, 6798, and videos 6795 and 6796.
Related to images 6791, 6792, 6794, 6797, 6798, and videos 6795 and 6796.
Alaina Willet, Kathy Gould’s lab, Vanderbilt University.
View Media
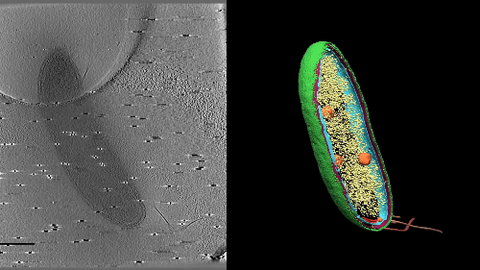
6569: Cryo-electron tomography of a Caulobacter bacterium
6569: Cryo-electron tomography of a Caulobacter bacterium
3D image of Caulobacter bacterium with various components highlighted: cell membranes (red and blue), protein shell (green), protein factories known as ribosomes (yellow), and storage granules (orange).
Peter Dahlberg, Stanford University.
View Media
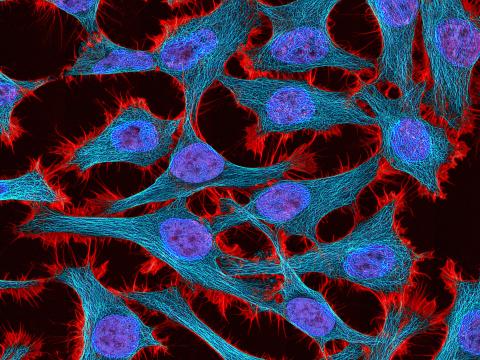
3521: HeLa cells
3521: HeLa cells
Multiphoton fluorescence image of HeLa cells stained with the actin binding toxin phalloidin (red), microtubules (cyan) and cell nuclei (blue). Nikon RTS2000MP custom laser scanning microscope. See related images 3518, 3519, 3520, 3522.
National Center for Microscopy and Imaging Research (NCMIR)
View Media
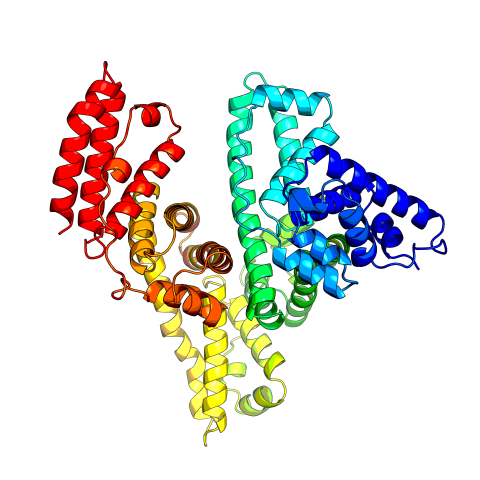
3745: Serum albumin structure 2
3745: Serum albumin structure 2
Serum albumin (SA) is the most abundant protein in the blood plasma of mammals. SA has a characteristic heart-shape structure and is a highly versatile protein. It helps maintain normal water levels in our tissues and carries almost half of all calcium ions in human blood. SA also transports some hormones, nutrients and metals throughout the bloodstream. Despite being very similar to our own SA, those from other animals can cause some mild allergies in people. Therefore, some scientists study SAs from humans and other mammals to learn more about what subtle structural or other differences cause immune responses in the body.
Related to entries 3744 and 3746
Related to entries 3744 and 3746
Wladek Minor, University of Virginia
View Media
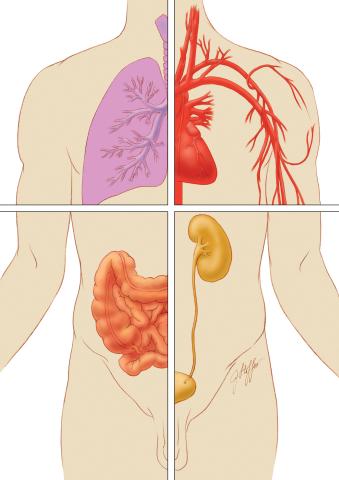
1280: Quartered torso
1280: Quartered torso
Cells function within organs and tissues, such as the lungs, heart, intestines, and kidney.
Judith Stoffer
View Media
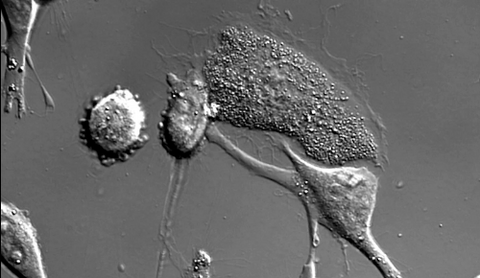
6966: Dying melanoma cells
6966: Dying melanoma cells
Melanoma (skin cancer) cells undergoing programmed cell death, also called apoptosis. This process was triggered by raising the pH of the medium that the cells were growing in. Melanoma in people cannot be treated by raising pH because that would also kill healthy cells. This video was taken using a differential interference contrast (DIC) microscope.
Dylan T. Burnette, Vanderbilt University School of Medicine.
View Media
6777: Human endoplasmic reticulum membrane protein complex
6777: Human endoplasmic reticulum membrane protein complex
A 3D model of the human endoplasmic reticulum membrane protein complex (EMC) that identifies its nine essential subunits. The EMC plays an important role in making membrane proteins, which are essential for all cellular processes. This is the first atomic-level depiction of the EMC. Its structure was obtained using single-particle cryo-electron microscopy.
Rebecca Voorhees, California Institute of Technology.
View Media
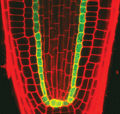
2329: Planting roots
2329: Planting roots
At the root tips of the mustard plant Arabidopsis thaliana (red), two proteins work together to control the uptake of water and nutrients. When the cell division-promoting protein called Short-root moves from the center of the tip outward, it triggers the production of another protein (green) that confines Short-root to the nutrient-filtering endodermis. The mechanism sheds light on how genes and proteins interact in a model organism and also could inform the engineering of plants.
Philip Benfey, Duke University
View Media
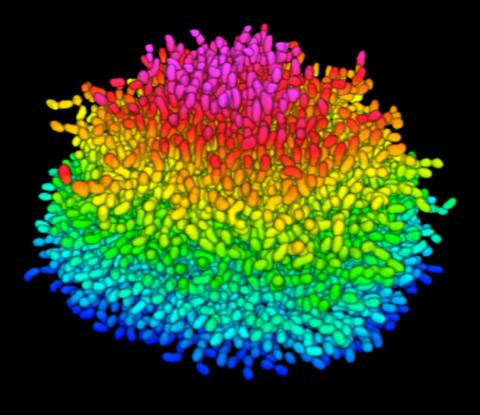
5825: A Growing Bacterial Biofilm
5825: A Growing Bacterial Biofilm
A growing Vibrio cholerae (cholera) biofilm. Cholera bacteria form colonies called biofilms that enable them to resist antibiotic therapy within the body and other challenges to their growth.
Each slightly curved comma shape represents an individual bacterium from assembled confocal microscopy images. Different colors show each bacterium’s position in the biofilm in relation to the surface on which the film is growing.
Each slightly curved comma shape represents an individual bacterium from assembled confocal microscopy images. Different colors show each bacterium’s position in the biofilm in relation to the surface on which the film is growing.
Jing Yan, Ph.D., and Bonnie Bassler, Ph.D., Department of Molecular Biology, Princeton University, Princeton, NJ.
View Media
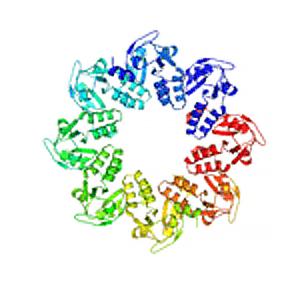
2377: Protein involved in cell division from Mycoplasma pneumoniae
2377: Protein involved in cell division from Mycoplasma pneumoniae
Model of a protein involved in cell division from Mycoplasma pneumoniae. This model, based on X-ray crystallography, revealed a structural domain not seen before. The protein is thought to be involved in cell division and cell wall biosynthesis.
Berkeley Structural Genomics Center, PSI
View Media
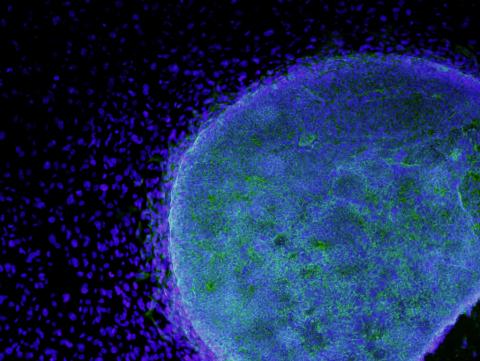
3269: Colony of human ES cells
3269: Colony of human ES cells
A colony of human embryonic stem cells (light blue) grows on fibroblasts (dark blue).
California Institute for Regenerative Medicine
View Media
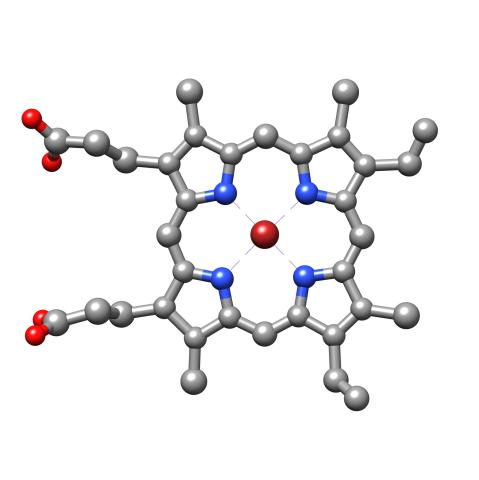
3539: Structure of heme, top view
3539: Structure of heme, top view
Molecular model of the struture of heme. Heme is a small, flat molecule with an iron ion (dark red) at its center. Heme is an essential component of hemoglobin, the protein in blood that carries oxygen throughout our bodies. This image first appeared in the September 2013 issue of Findings Magazine. View side view of heme here 3540.
Rachel Kramer Green, RCSB Protein Data Bank
View Media
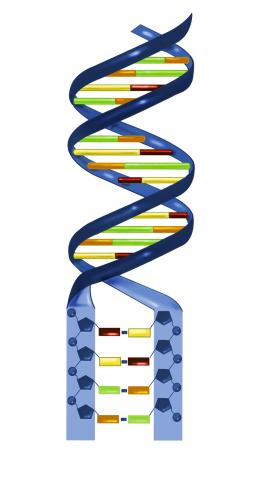
2541: Nucleotides make up DNA
2541: Nucleotides make up DNA
DNA consists of two long, twisted chains made up of nucleotides. Each nucleotide contains one base, one phosphate molecule, and the sugar molecule deoxyribose. The bases in DNA nucleotides are adenine, thymine, cytosine, and guanine. See image 2542 for a labeled version of this illustration. Featured in The New Genetics.
Crabtree + Company
View Media
5752: Genetically identical mycobacteria respond differently to antibiotic 2
5752: Genetically identical mycobacteria respond differently to antibiotic 2
Antibiotic resistance in microbes is a serious health concern. So researchers have turned their attention to how bacteria undo the action of some antibiotics. Here, scientists set out to find the conditions that help individual bacterial cells survive in the presence of the antibiotic rifampicin. The research team used Mycobacterium smegmatis, a more harmless relative of Mycobacterium tuberculosis, which infects the lung and other organs to cause serious disease.
In this video, genetically identical mycobacteria are growing in a miniature growth chamber called a microfluidic chamber. Using live imaging, the researchers found that individual mycobacteria will respond differently to the antibiotic, depending on the growth stage and other timing factors. The researchers used genetic tagging with green fluorescent protein to distinguish cells that can resist rifampicin and those that cannot. With this gene tag, cells tolerant of the antibiotic light up in green and those that are susceptible in violet, enabling the team to monitor the cells' responses in real time.
To learn more about how the researchers studied antibiotic resistance in mycobacteria, see this news release from Tufts University. Related to image 5751.
In this video, genetically identical mycobacteria are growing in a miniature growth chamber called a microfluidic chamber. Using live imaging, the researchers found that individual mycobacteria will respond differently to the antibiotic, depending on the growth stage and other timing factors. The researchers used genetic tagging with green fluorescent protein to distinguish cells that can resist rifampicin and those that cannot. With this gene tag, cells tolerant of the antibiotic light up in green and those that are susceptible in violet, enabling the team to monitor the cells' responses in real time.
To learn more about how the researchers studied antibiotic resistance in mycobacteria, see this news release from Tufts University. Related to image 5751.
Bree Aldridge, Tufts University
View Media
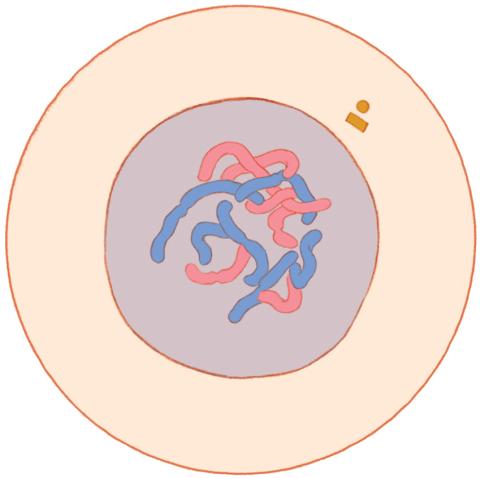
1316: Mitosis - interphase
1316: Mitosis - interphase
A cell in interphase, at the start of mitosis: Chromosomes duplicate, and the copies remain attached to each other. Mitosis is responsible for growth and development, as well as for replacing injured or worn out cells throughout the body. For simplicity, mitosis is illustrated here with only six chromosomes.
Judith Stoffer
View Media
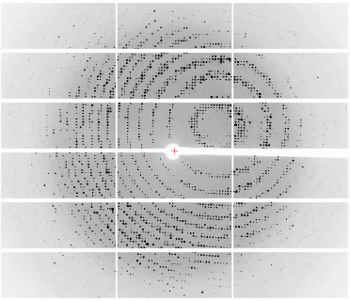
6765: X-ray diffraction pattern from a crystallized cefotaxime-CCD-1 complex
6765: X-ray diffraction pattern from a crystallized cefotaxime-CCD-1 complex
CCD-1 is an enzyme produced by the bacterium Clostridioides difficile that helps it resist antibiotics. Researchers crystallized complexes where a CCD-1 molecule and a molecule of the antibiotic cefotaxime were bound together. Then, they shot X-rays at the complexes to determine their structure—a process known as X-ray crystallography. This image shows the X-ray diffraction pattern of a complex.
Related to images 6764, 6766, and 6767.
Related to images 6764, 6766, and 6767.
Keith Hodgson, Stanford University.
View Media
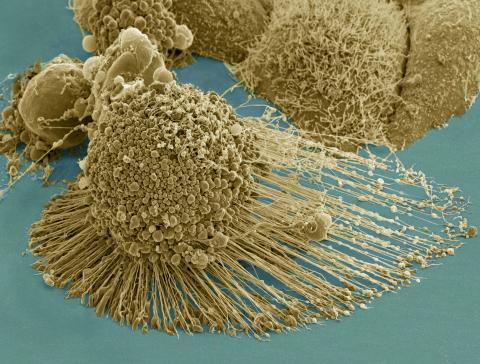
3519: HeLa cells
3519: HeLa cells
Scanning electron micrograph of an apoptotic HeLa cell. Zeiss Merlin HR-SEM. See related images 3518, 3520, 3521, 3522.
National Center for Microscopy and Imaging Research
View Media
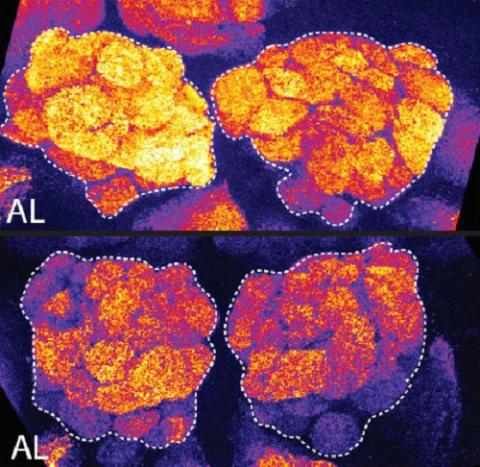
3490: Brains of sleep-deprived and well-rested fruit flies
3490: Brains of sleep-deprived and well-rested fruit flies
On top, the brain of a sleep-deprived fly glows orange because of Bruchpilot, a communication protein between brain cells. These bright orange brain areas are associated with learning. On the bottom, a well-rested fly shows lower levels of Bruchpilot, which might make the fly ready to learn after a good night's rest.
Chiara Cirelli, University of Wisconsin-Madison
View Media
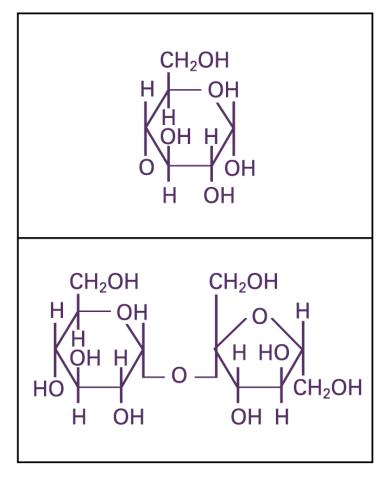
2500: Glucose and sucrose
2500: Glucose and sucrose
Glucose (top) and sucrose (bottom) are sugars made of carbon, hydrogen, and oxygen atoms. Carbohydrates include simple sugars like these and are the main source of energy for the human body.
Crabtree + Company
View Media
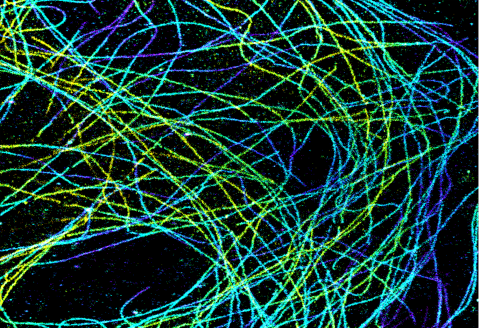
6891: Microtubules in African green monkey cells
6891: Microtubules in African green monkey cells
Microtubules in African green monkey cells. Microtubules are strong, hollow fibers that provide cells with structural support. Here, the microtubules have been color-coded based on their distance from the microscope lens: purple is closest to the lens, and yellow is farthest away. This image was captured using Stochastic Optical Reconstruction Microscopy (STORM).
Related to images 6889, 6890, and 6892.
Related to images 6889, 6890, and 6892.
Melike Lakadamyali, Perelman School of Medicine at the University of Pennsylvania.
View Media
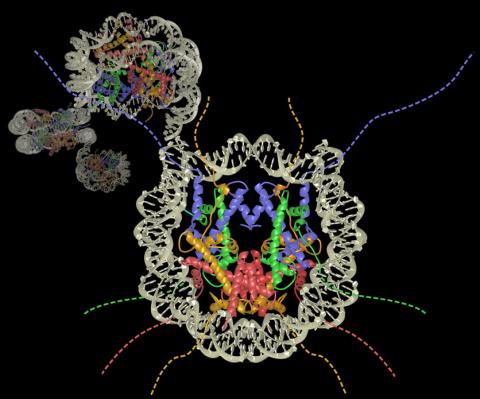
2741: Nucleosome
2741: Nucleosome
Like a strand of white pearls, DNA wraps around an assembly of special proteins called histones (colored) to form the nucleosome, a structure responsible for regulating genes and condensing DNA strands to fit into the cell's nucleus. Researchers once thought that nucleosomes regulated gene activity through their histone tails (dotted lines), but a 2010 study revealed that the structures' core also plays a role. The finding sheds light on how gene expression is regulated and how abnormal gene regulation can lead to cancer.
Karolin Luger, Colorado State University
View Media
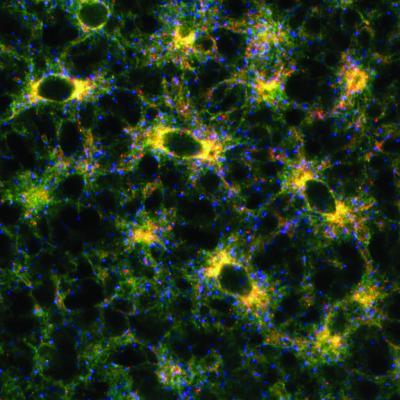
2649: Endoplasmic reticulum
2649: Endoplasmic reticulum
Fluorescent markers show the interconnected web of tubes and compartments in the endoplasmic reticulum. The protein atlastin helps build and maintain this critical part of cells. The image is from a July 2009 news release.
Andrea Daga, Eugenio Medea Scientific Institute (Conegliano, Italy)
View Media
3597: DNA replication origin recognition complex (ORC)
3597: DNA replication origin recognition complex (ORC)
A study published in March 2012 used cryo-electron microscopy to determine the structure of the DNA replication origin recognition complex (ORC), a semi-circular, protein complex (yellow) that recognizes and binds DNA to start the replication process. The ORC appears to wrap around and bend approximately 70 base pairs of double stranded DNA (red and blue). Also shown is the protein Cdc6 (green), which is also involved in the initiation of DNA replication. Related to video 3307 that shows the structure from different angles. From a Brookhaven National Laboratory news release, "Study Reveals How Protein Machinery Binds and Wraps DNA to Start Replication."
Huilin Li, Brookhaven National Laboratory
View Media
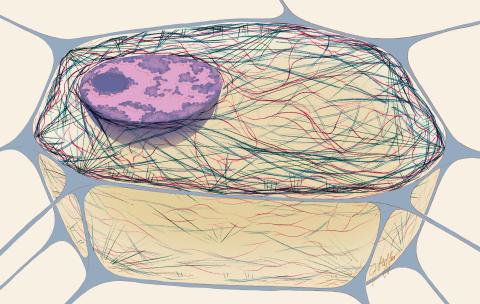
1272: Cytoskeleton
1272: Cytoskeleton
The three fibers of the cytoskeleton--microtubules in blue, intermediate filaments in red, and actin in green--play countless roles in the cell.
Judith Stoffer
View Media
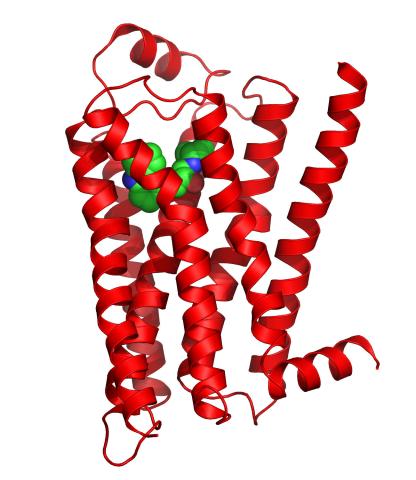
3358: Beta 2-adrenergic receptor
3358: Beta 2-adrenergic receptor
The receptor is shown bound to a partial inverse agonist, carazolol.
Raymond Stevens, The Scripps Research Institute
View Media
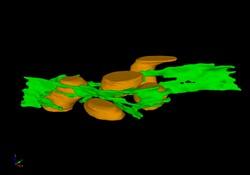
2635: Mitochondria and endoplasmic reticulum
2635: Mitochondria and endoplasmic reticulum
A computer model shows how the endoplasmic reticulum is close to and almost wraps around mitochondria in the cell. The endoplasmic reticulum is lime green and the mitochondria are yellow. This image relates to a July 27, 2009 article in Computing Life.
Bridget Wilson, University of New Mexico
View Media