Switch to List View
Image and Video Gallery
This is a searchable collection of scientific photos, illustrations, and videos. The images and videos in this gallery are licensed under Creative Commons Attribution Non-Commercial ShareAlike 3.0. This license lets you remix, tweak, and build upon this work non-commercially, as long as you credit and license your new creations under identical terms.
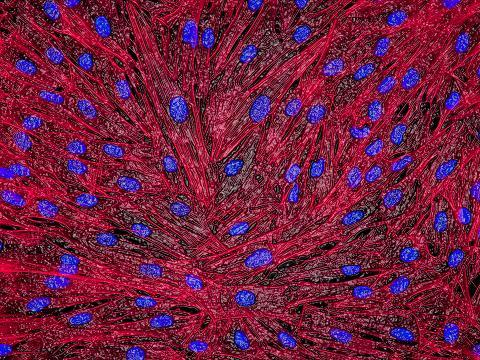
3670: DNA and actin in cultured fibroblast cells
3670: DNA and actin in cultured fibroblast cells
DNA (blue) and actin (red) in cultured fibroblast cells.
Tom Deerinck, National Center for Microscopy and Imaging Research (NCMIR)
View Media

2543: DNA replication illustration
2543: DNA replication illustration
During DNA replication, each strand of the original molecule acts as a template for the synthesis of a new, complementary DNA strand. See image 2544 for a labeled version of this illustration.
Crabtree + Company
View Media
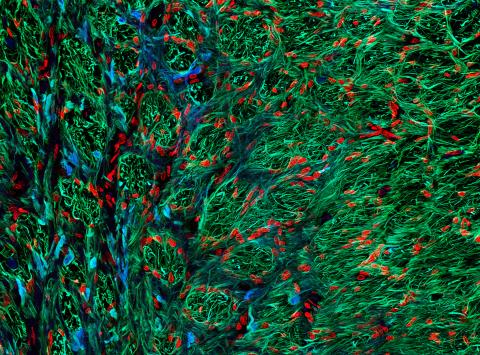
5852: Optic nerve astrocytes
5852: Optic nerve astrocytes
Astrocytes in the cross section of a human optic nerve head
Tom Deerinck and Keunyoung (“Christine”) Kim, NCMIR
View Media
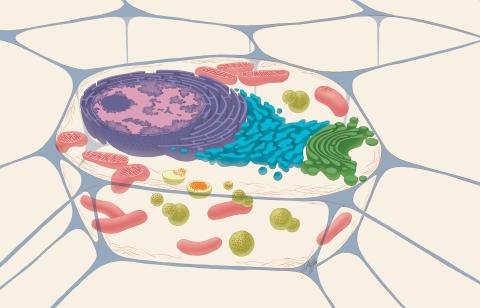
1274: Animal cell
1274: Animal cell
A typical animal cell, sliced open to reveal a cross-section of organelles.
Judith Stoffer
View Media
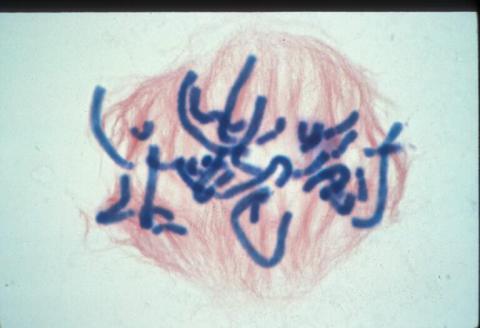
1016: Lily mitosis 06
1016: Lily mitosis 06
A light microscope image of a cell from the endosperm of an African globe lily (Scadoxus katherinae). This is one frame of a time-lapse sequence that shows cell division in action. The lily is considered a good organism for studying cell division because its chromosomes are much thicker and easier to see than human ones. Staining shows microtubules in red and chromosomes in blue. Here, condensed chromosomes are clearly visible and are starting to line up.
Related to images 1010, 1011, 1012, 1013, 1014, 1015, 1017, 1018, 1019, and 1021.
Related to images 1010, 1011, 1012, 1013, 1014, 1015, 1017, 1018, 1019, and 1021.
Andrew S. Bajer, University of Oregon, Eugene
View Media
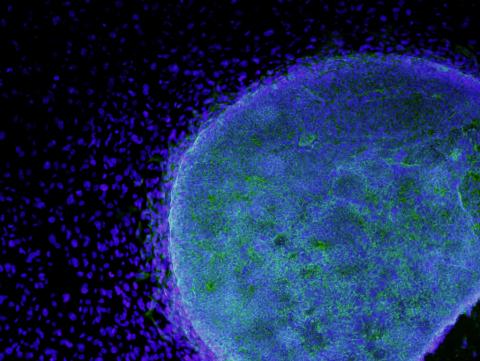
3269: Colony of human ES cells
3269: Colony of human ES cells
A colony of human embryonic stem cells (light blue) grows on fibroblasts (dark blue).
California Institute for Regenerative Medicine
View Media
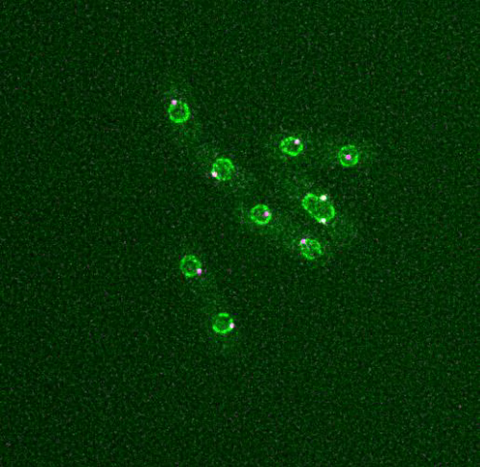
6795: Dividing yeast cells with nuclear envelopes and spindle pole bodies
6795: Dividing yeast cells with nuclear envelopes and spindle pole bodies
Time-lapse video of yeast cells undergoing cell division. Nuclear envelopes are shown in green, and spindle pole bodies, which help pull apart copied genetic information, are shown in magenta. This video was captured using wide-field microscopy with deconvolution.
Related to images 6791, 6792, 6793, 6794, 6797, 6798, and video 6796.
Related to images 6791, 6792, 6793, 6794, 6797, 6798, and video 6796.
Alaina Willet, Kathy Gould’s lab, Vanderbilt University.
View Media
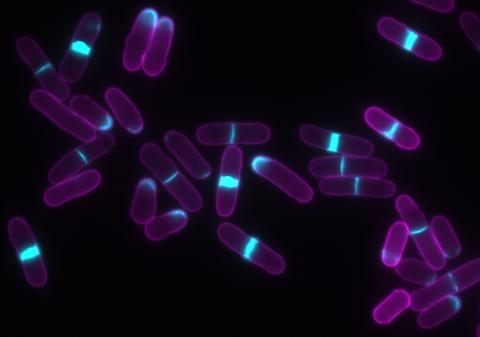
6797: Yeast cells with accumulated cell wall material
6797: Yeast cells with accumulated cell wall material
Yeast cells that abnormally accumulate cell wall material (blue) at their ends and, when preparing to divide, in their middles. This image was captured using wide-field microscopy with deconvolution.
Related to images 6791, 6792, 6793, 6794, 6798, and videos 6795 and 6796.
Related to images 6791, 6792, 6793, 6794, 6798, and videos 6795 and 6796.
Alaina Willet, Kathy Gould’s lab, Vanderbilt University.
View Media
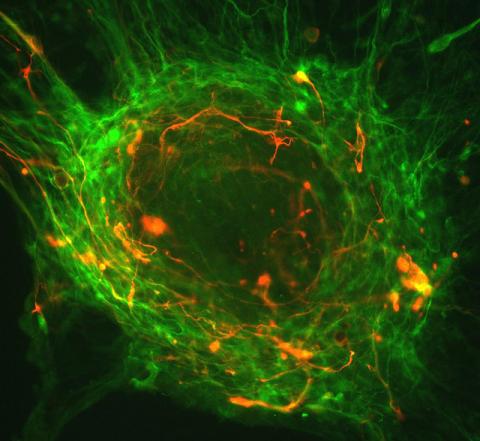
3276: Human ES cells differentiating into neurons
3276: Human ES cells differentiating into neurons
This image shows hundreds of human embryonic stem cells in various stages of differentiating into neurons. Some cells have become neurons (red), while others are still precursors of nerve cells (green). The yellow is an imaging artifact resulting when cells in both stages are on top of each other. Image and caption information courtesy of the California Institute for Regenerative Medicine.
Guoping Fan lab, University of California, Los Angeles, via CIRM
View Media

6344: Drosophila
6344: Drosophila
Two adult fruit flies (Drosophila)
Dr. Vicki Losick, MDI Biological Laboratory, www.mdibl.org
View Media

3423: White Poppy (cropped)
3423: White Poppy (cropped)
A cropped image of a white poppy. View poppy uncropped here 3424.
Judy Coyle, Donald Danforth Plant Science Center
View Media
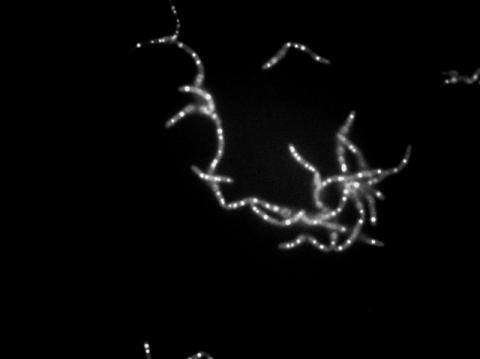
3481: Bacillus anthracis being killed
3481: Bacillus anthracis being killed
Bacillus anthracis (anthrax) cells being killed by a fluorescent trans-translation inhibitor, which disrupts bacterial protein synthesis. The inhibitor is naturally fluorescent and looks blue when it is excited by ultraviolet light in the microscope. This is a black-and-white version of Image 3525.
John Alumasa, Keiler Laboratory, Pennsylvania State University
View Media
2796: Anti-tumor drug ecteinascidin 743 (ET-743), structure without hydrogens 03
2796: Anti-tumor drug ecteinascidin 743 (ET-743), structure without hydrogens 03
Ecteinascidin 743 (ET-743, brand name Yondelis), was discovered and isolated from a sea squirt, Ecteinascidia turbinata, by NIGMS grantee Kenneth Rinehart at the University of Illinois. It was synthesized by NIGMS grantees E.J. Corey and later by Samuel Danishefsky. Multiple versions of this structure are available as entries 2790-2797.
Timothy Jamison, Massachusetts Institute of Technology
View Media

3755: Cryo-EM reveals how the HIV capsid attaches to a human protein to evade immune detection
3755: Cryo-EM reveals how the HIV capsid attaches to a human protein to evade immune detection
The illustration shows the capsid of human immunodeficiency virus (HIV) whose molecular features were resolved with cryo-electron microscopy (cryo-EM). On the left, the HIV capsid is "naked," a state in which it would be easily detected by and removed from cells. However, as shown on the right, when the viral capsid binds to and is covered with a host protein, called cyclophilin A (shown in red), it evades detection and enters and invades the human cell to use it to establish an infection. To learn more about how cyclophilin A helps HIV infect cells and how scientists used cryo-EM to find out the mechanism by which the HIV capsid attaches to cyclophilin A, see this news release by the University of Illinois. A study reporting these findings was published in the journal Nature Communications.
Juan R. Perilla, University of Illinois at Urbana-Champaign
View Media
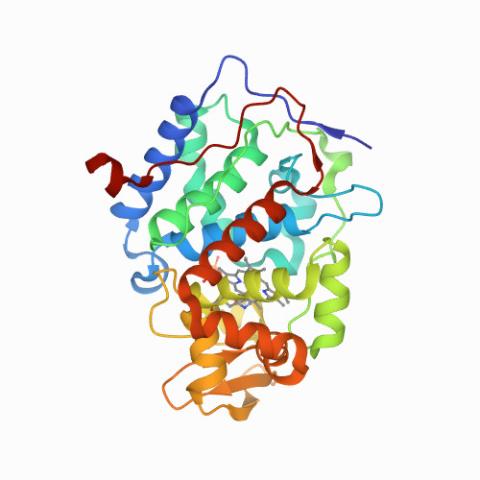
6762: CCP enzyme
6762: CCP enzyme
The enzyme CCP is found in the mitochondria of baker’s yeast. Scientists study the chemical reactions that CCP triggers, which involve a water molecule, iron, and oxygen. This structure was determined using an X-ray free electron laser.
Protein Data Bank.
View Media
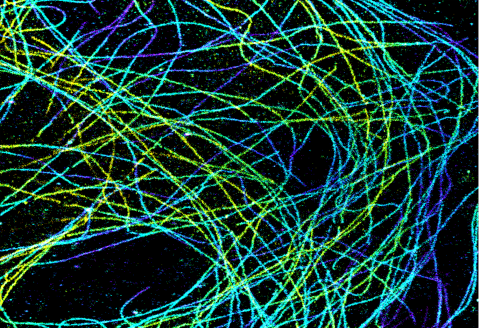
6891: Microtubules in African green monkey cells
6891: Microtubules in African green monkey cells
Microtubules in African green monkey cells. Microtubules are strong, hollow fibers that provide cells with structural support. Here, the microtubules have been color-coded based on their distance from the microscope lens: purple is closest to the lens, and yellow is farthest away. This image was captured using Stochastic Optical Reconstruction Microscopy (STORM).
Related to images 6889, 6890, and 6892.
Related to images 6889, 6890, and 6892.
Melike Lakadamyali, Perelman School of Medicine at the University of Pennsylvania.
View Media

2319: Mapping metabolic activity
2319: Mapping metabolic activity
Like a map showing heavily traveled roads, this mathematical model of metabolic activity inside an E. coli cell shows the busiest pathway in white. Reaction pathways used less frequently by the cell are marked in red (moderate activity) and green (even less activity). Visualizations like this one may help scientists identify drug targets that block key metabolic pathways in bacteria.
Albert-László Barabási, University of Notre Dame
View Media
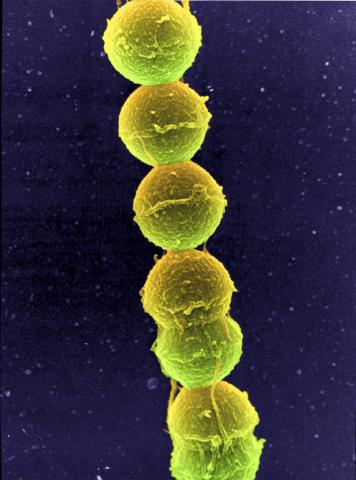
1157: Streptococcus bacteria
1157: Streptococcus bacteria
Image of Streptococcus, a type (genus) of spherical bacteria that can colonize the throat and back of the mouth. Stroptococci often occur in pairs or in chains, as shown here.
Tina Weatherby Carvalho, University of Hawaii at Manoa
View Media
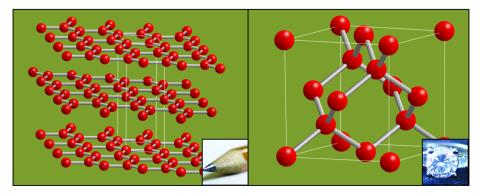
2507: Carbon building blocks (with examples)
2507: Carbon building blocks (with examples)
The arrangement of identical molecular components can make a dramatic difference. For example, carbon atoms can be arranged into dull graphite (left) or sparkly diamonds (right). See image 2506 for an illustration without examples.
Crabtree + Company
View Media
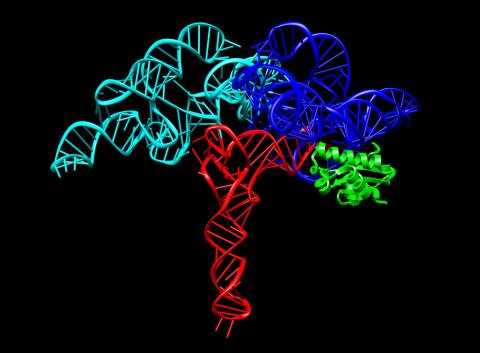
3660: Ribonuclease P structure
3660: Ribonuclease P structure
Ribbon diagram showing the structure of Ribonuclease P with tRNA.
PDB entry 3Q1Q, molecular modeling by Fred Friedman, NIGMS
View Media
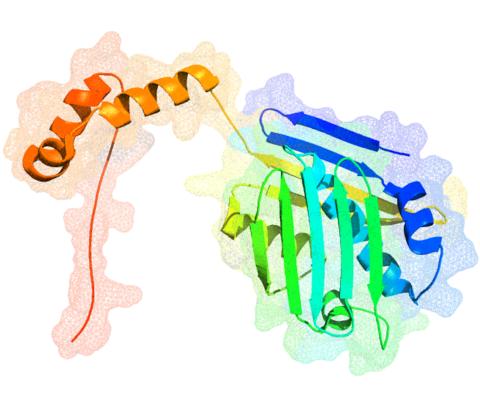
3354: Hsp33 figure 1
3354: Hsp33 figure 1
Featured in the March 15, 2012 issue of Biomedical Beat. Related to Hsp33 Figure 2, image 3355.
Ursula Jakob and Dana Reichmann, University of Michigan
View Media
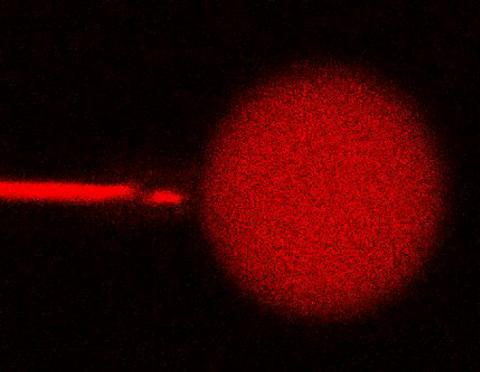
3583: Bee venom toxin destroying a cell
3583: Bee venom toxin destroying a cell
This video condenses 6.5 minutes into less than a minute to show how the toxin in bee venom, called melittin, destroys an animal or bacterial cell. What looks like a red balloon is an artificial cell filled with red dye. Melittin molecules are colored green and float on the cell's surface like twigs on a pond. As melittin accumulates on the cell's membrane, the membrane expands to accommodate it. In the video, the membrane stretches into a column on the left. When melittin levels reach a critical threshold, countless pinhole leaks burst open in the membrane. The cell's vital fluids (red dye in the video) leak out through these pores. Within minutes, the cell collapses.
Huey Huang, Rice University
View Media
1086: Natcher Building 06
1086: Natcher Building 06
NIGMS staff are located in the Natcher Building on the NIH campus.
Alisa Machalek, National Institute of General Medical Sciences
View Media
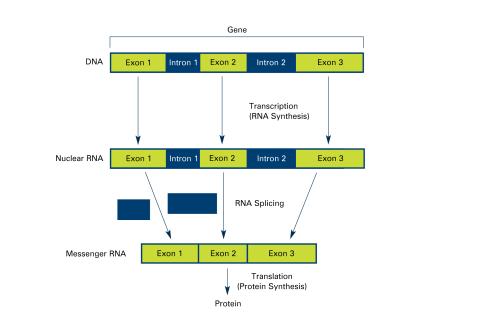
2551: Introns (with labels)
2551: Introns (with labels)
Genes are often interrupted by stretches of DNA (introns, blue) that do not contain instructions for making a protein. The DNA segments that do contain protein-making instructions are known as exons (green). See image 2550 for an unlabeled version of this illustration. Featured in The New Genetics.
Crabtree + Company
View Media
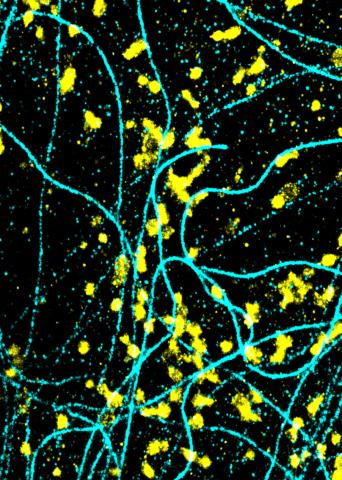
6889: Lysosomes and microtubules
6889: Lysosomes and microtubules
Lysosomes (yellow) and detyrosinated microtubules (light blue). Lysosomes are bubblelike organelles that take in molecules and use enzymes to break them down. Microtubules are strong, hollow fibers that provide structural support to cells. The researchers who took this image found that in epithelial cells, detyrosinated microtubules are a small subset of fibers, and they concentrate lysosomes around themselves. This image was captured using Stochastic Optical Reconstruction Microscopy (STORM).
Related to images 6890, 6891, and 6892.
Related to images 6890, 6891, and 6892.
Melike Lakadamyali, Perelman School of Medicine at the University of Pennsylvania.
View Media
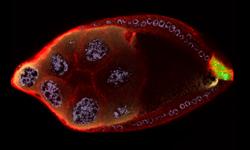
2309: Cellular polarity
2309: Cellular polarity
As an egg cell develops, a process called polarization controls what parts ultimately become the embryo's head and tail. This picture shows an egg of the fruit fly Drosophila. Red and green mark two types of signaling proteins involved in polarization. Disrupting these signals can scramble the body plan of the embryo, leading to severe developmental disorders.
Wu-Min Deng, Florida State University
View Media
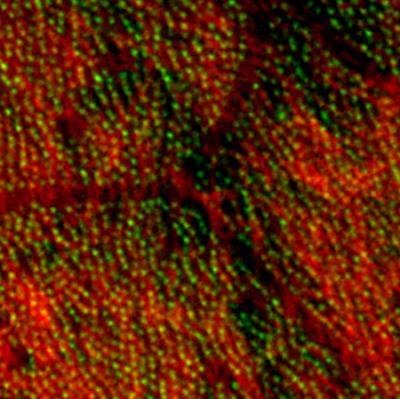
3292: Centrioles anchor cilia in planaria
3292: Centrioles anchor cilia in planaria
Centrioles (green) anchor cilia (red), which project on the surface of pharynx cells of the freshwater planarian Schmidtea mediterranea. Centrioles require cellular structures called centrosomes for assembly in other animal species, but this flatworm known for its regenerative ability was unexpectedly found to lack centrosomes. From a Stowers University news release.
Juliette Azimzadeh, University of California, San Francisco
View Media
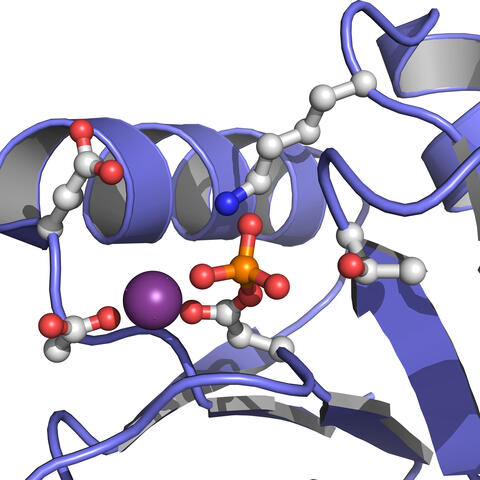
3412: Active Site of E. coli response regulator PhoB
3412: Active Site of E. coli response regulator PhoB
Active site of E. coli response regulator PhoB.
Ann Stock, Rutgers University
View Media

2414: Pig trypsin (3)
2414: Pig trypsin (3)
Crystals of porcine trypsin protein created for X-ray crystallography, which can reveal detailed, three-dimensional protein structures.
Alex McPherson, University of California, Irvine
View Media
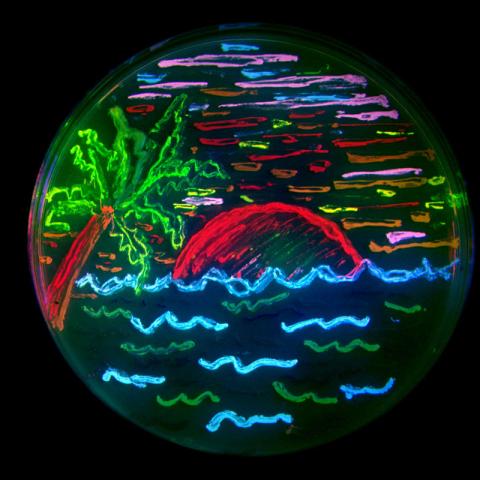
3492: Glowing bacteria make a pretty postcard
3492: Glowing bacteria make a pretty postcard
This tropical scene, reminiscent of a postcard from Key West, is actually a petri dish containing an artistic arrangement of genetically engineered bacteria. The image showcases eight of the fluorescent proteins created in the laboratory of the late Roger Y. Tsien, a cell biologist at the University of California, San Diego. Tsien, along with Osamu Shimomura of the Marine Biology Laboratory and Martin Chalfie of Columbia University, share the 2008 Nobel Prize in chemistry for their work on green fluorescent protein-a naturally glowing molecule from jellyfish that has become a powerful tool for studying molecules inside living cells.
Nathan C. Shaner, The Scintillon Institute
View Media
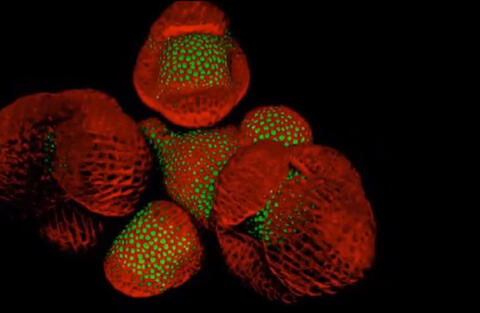
6503: Arabidopsis Thaliana: Flowers Spring to Life
6503: Arabidopsis Thaliana: Flowers Spring to Life
This image capture shows how a single gene, STM, plays a starring role in plant development. This gene acts like a molecular fountain of youth, keeping cells ever-young until it’s time to grow up and commit to making flowers and other plant parts. Because of its ease of use and low cost, Arabidopsis is a favorite model for scientists to learn the basic principles driving tissue growth and regrowth for humans as well as the beautiful plants outside your window. Image captured from video Watch Flowers Spring to Life, featured in the NIH Director's Blog: Watch Flowers Spring to Life.
Nathanaёl Prunet NIH Support: National Institute of General Medical Sciences
View Media
6850: Himastatin and bacteria
6850: Himastatin and bacteria
A model of the molecule himastatin overlaid on an image of Bacillus subtilis bacteria. Scientists first isolated himastatin from the bacterium Streptomyces himastatinicus, and the molecule shows antibiotic activity. The researchers who created this image developed a new, more concise way to synthesize himastatin so it can be studied more easily. They also tested the effects of himastatin and derivatives of the molecule on B. subtilis.
More information about the research that produced this image can be found in the Science paper “Total synthesis of himastatin” by D’Angelo et al.
Related to image 6848 and video 6851.
More information about the research that produced this image can be found in the Science paper “Total synthesis of himastatin” by D’Angelo et al.
Related to image 6848 and video 6851.
Mohammad Movassaghi, Massachusetts Institute of Technology.
View Media
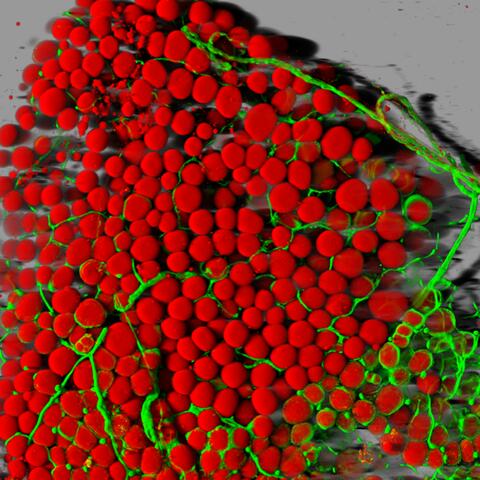
3600: Fat cells (red) and blood vessels (green)
3600: Fat cells (red) and blood vessels (green)
A mouse's fat cells (red) are shown surrounded by a network of blood vessels (green). Fat cells store and release energy, protect organs and nerve tissues, insulate us from the cold, and help us absorb important vitamins.
This image was part of the Life: Magnified exhibit that ran from June 3, 2014, to January 21, 2015, at Dulles International Airport.
This image was part of the Life: Magnified exhibit that ran from June 3, 2014, to January 21, 2015, at Dulles International Airport.
Daniela Malide, National Heart, Lung, and Blood Institute, National Institutes of Health
View Media
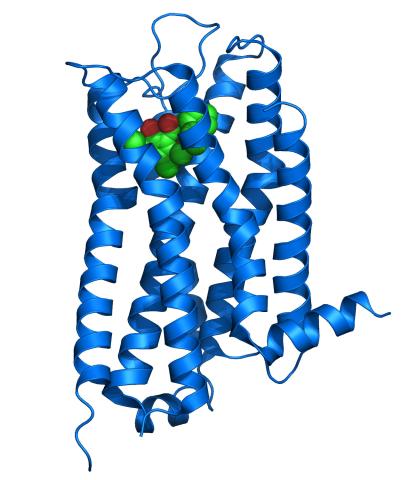
3363: Dopamine D3 receptor
3363: Dopamine D3 receptor
The receptor is shown bound to an antagonist, eticlopride
Raymond Stevens, The Scripps Research Institute
View Media
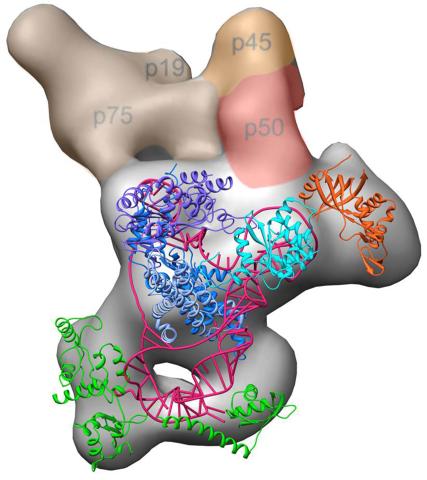
3459: Structure of telomerase
3459: Structure of telomerase
Scientists recently discovered the full molecular structure of telomerase, an enzyme important to aging and cancer. Within each cell, telomerase maintains the telomeres, or end pieces, of a chromosome, preserving genetic data and extending the life of the cell. In their study, a team from UCLA and UC Berkeley found the subunit p50, shown in red, to be a keystone in the enzyme's structure and function. Featured in the May 16, 2013 issue of Biomedical Beat.
Jiansen Jiang, Edward J. Miracco, Z. Hong Zhou and Juli Feigon, University of California, Los Angeles; Kathleen Collins, University of California, Berkeley
View Media
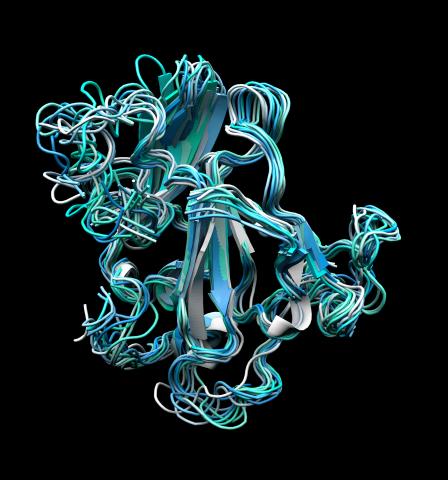
5866: Structure of a key antigen protein involved with Hepatitis C Virus infection
5866: Structure of a key antigen protein involved with Hepatitis C Virus infection
A three-dimensional representation of the structure of E2, a key antigen protein involved with hepatitis C virus infection.
Mansun Law Associate Professor Department of Immunolgy and Microbial Science The Scripps Research Institute
View Media
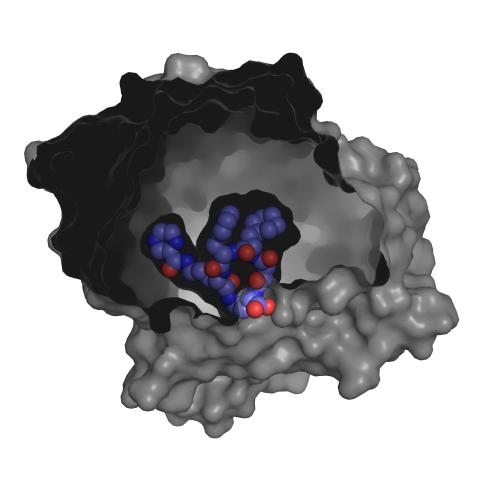
3415: X-ray co-crystal structure of Src kinase bound to a DNA-templated macrocycle inhibitor 3
3415: X-ray co-crystal structure of Src kinase bound to a DNA-templated macrocycle inhibitor 3
X-ray co-crystal structure of Src kinase bound to a DNA-templated macrocycle inhibitor. Related to 3413, 3414, 3416, 3417, 3418, and 3419.
Markus A. Seeliger, Stony Brook University Medical School and David R. Liu, Harvard University
View Media
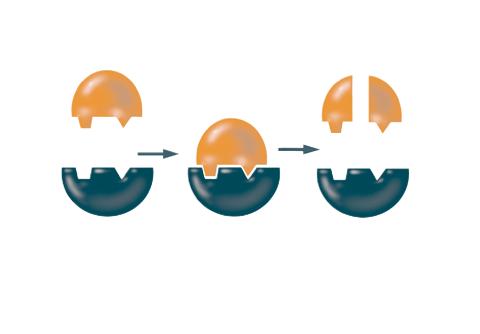
2521: Enzymes convert subtrates into products
2521: Enzymes convert subtrates into products
Enzymes convert substrates into products very quickly. See image 2522 for a labeled version of this illustration. Featured in The Chemistry of Health.
Crabtree + Company
View Media
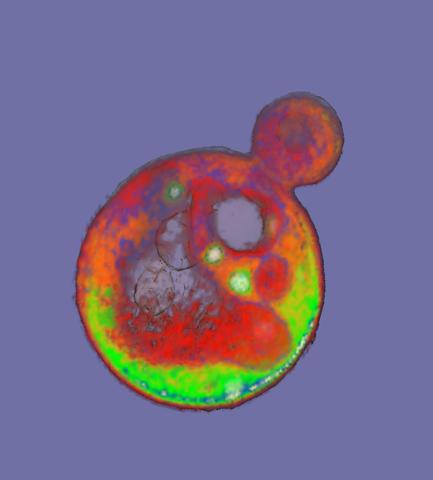
2307: Cells frozen in time
2307: Cells frozen in time
The fledgling field of X-ray microscopy lets researchers look inside whole cells rapidly frozen to capture their actions at that very moment. Here, a yeast cell buds before dividing into two. Colors show different parts of the cell. Seeing whole cells frozen in time will help scientists observe cells' complex structures and follow how molecules move inside them.
Carolyn Larabell, University of California, San Francisco, and the Lawrence Berkeley National Laboratory
View Media
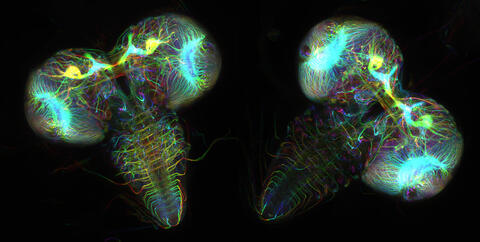
6808: Fruit fly larvae brains showing tubulin
6808: Fruit fly larvae brains showing tubulin
Two fruit fly (Drosophila melanogaster) larvae brains with neurons expressing fluorescently tagged tubulin protein. Tubulin makes up strong, hollow fibers called microtubules that play important roles in neuron growth and migration during brain development. This image was captured using confocal microscopy, and the color indicates the position of the neurons within the brain.
Vladimir I. Gelfand, Feinberg School of Medicine, Northwestern University.
View Media
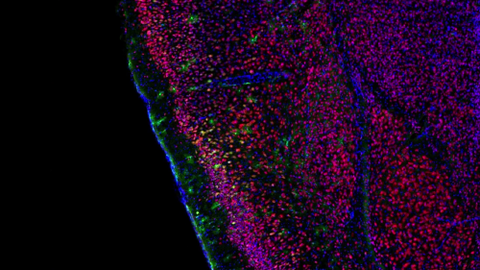
6781: Video of Calling Cards in a mouse brain
6781: Video of Calling Cards in a mouse brain
The green spots in this mouse brain are cells labeled with Calling Cards, a technology that records molecular events in brain cells as they mature. Understanding these processes during healthy development can guide further research into what goes wrong in cases of neuropsychiatric disorders. Also fluorescently labeled in this video are neurons (red) and nuclei (blue). Calling Cards and its application are described in the Cell paper “Self-Reporting Transposons Enable Simultaneous Readout of Gene Expression and Transcription Factor Binding in Single Cells” by Moudgil et al.; and the Proceedings of the National Academy of Sciences paper “A viral toolkit for recording transcription factor–DNA interactions in live mouse tissues” by Cammack et al. This video was created for the NIH Director’s Blog post The Amazing Brain: Tracking Molecular Events with Calling Cards.
Related to image 6780.
Related to image 6780.
NIH Director's Blog
View Media
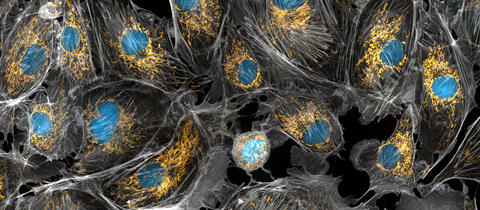
5762: Panorama view of golden mitochondria
5762: Panorama view of golden mitochondria
Mitochondria are the powerhouses of the cells, generating the energy the cells need to do their tasks and to stay alive. Researchers have studied mitochondria for some time because when these cell organelles don't work as well as they should, several diseases develop. In this photograph of cow cells taken with a microscope, the mitochondria were stained in bright yellow to visualize them in the cell. The large blue dots are the cell nuclei and the gray web is the cytoskeleton of the cells.
Torsten Wittmann, University of California, San Francisco
View Media

3327: Diversity oriented synthesis: generating skeletal diversity using folding processes
3327: Diversity oriented synthesis: generating skeletal diversity using folding processes
This 1 1/2-minute video animation was produced for chemical biologist Stuart Schreiber's lab page. The animation shows how diverse chemical structures can be produced in the lab.
Eric Keller
View Media
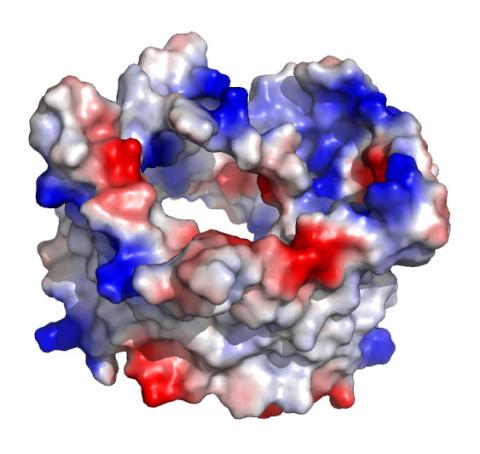
2488: VDAC-1 (1)
2488: VDAC-1 (1)
The structure of the pore-forming protein VDAC-1 from humans. This molecule mediates the flow of products needed for metabolism--in particular the export of ATP--across the outer membrane of mitochondria, the power plants for eukaryotic cells. VDAC-1 is involved in metabolism and the self-destruction of cells--two biological processes central to health.
Related to images 2491, 2494, and 2495.
Related to images 2491, 2494, and 2495.
Gerhard Wagner, Harvard Medical School
View Media
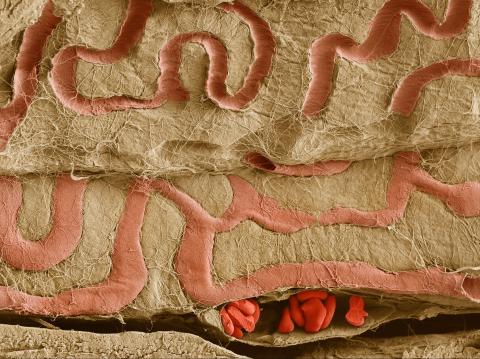
3739: Scanning electron microscopy of the ECM on the surface of a calf muscle
3739: Scanning electron microscopy of the ECM on the surface of a calf muscle
This image shows the extracellular matrix (ECM) on the surface of a soleus (lower calf) muscle in light brown and blood vessels in pink. Near the bottom of the photo, a vessel is opened up to reveal red blood cells. Scientists know less about the ECM in muscle than in other tissues, but it's increasingly clear that the ECM is critical to muscle function, and disruption of the ECM has been associated with many muscle disorders. The ECM in muscles stores and releases growth factors, suggesting that it might play a role in cellular communication.
Tom Deerinck, National Center for Microscopy and Imaging Research (NCMIR)
View Media
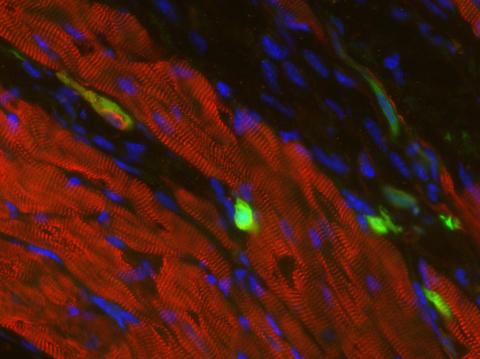
3273: Heart muscle with reprogrammed skin cells
3273: Heart muscle with reprogrammed skin cells
Skins cells were reprogrammed into heart muscle cells. The cells highlighted in green are remaining skin cells. Red indicates a protein that is unique to heart muscle. The technique used to reprogram the skin cells into heart cells could one day be used to mend heart muscle damaged by disease or heart attack. Image and caption information courtesy of the California Institute for Regenerative Medicine.
Deepak Srivastava, Gladstone Institute of Cardiovascular Disease, via CIRM
View Media
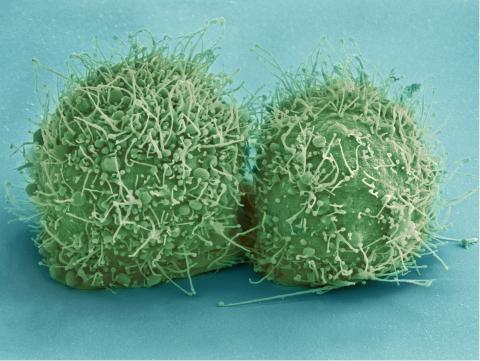
3518: HeLa cells
3518: HeLa cells
Scanning electron micrograph of just-divided HeLa cells. Zeiss Merlin HR-SEM. See related images 3519, 3520, 3521, 3522.
National Center for Microscopy and Imaging Research
View Media
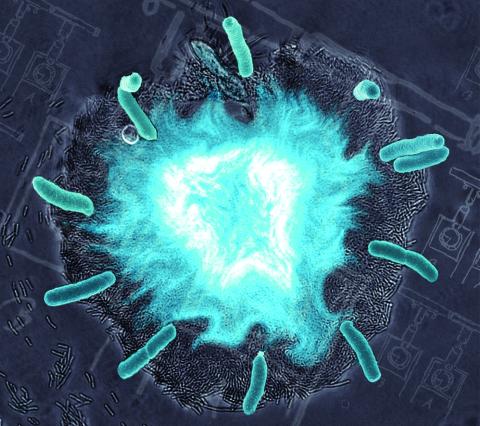
2725: Supernova bacteria
2725: Supernova bacteria
Bacteria engineered to act as genetic clocks flash in synchrony. Here, a "supernova" burst in a colony of coupled genetic clocks just after reaching critical cell density. Superimposed: A diagram from the notebook of Christiaan Huygens, who first characterized synchronized oscillators in the 17th century.
Jeff Hasty, UCSD
View Media
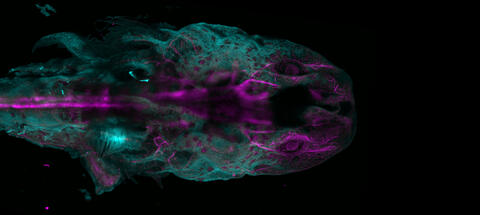
6927: Axolotl showing nervous system
6927: Axolotl showing nervous system
The head of an axolotl—a type of salamander—that has been genetically modified so that its developing nervous system glows purple and its Schwann cell nuclei appear light blue. Schwann cells insulate and provide nutrients to peripheral nerve cells. Researchers often study axolotls for their extensive regenerative abilities. They can regrow tails, limbs, spinal cords, brains, and more. The researcher who took this image focuses on the role of the peripheral nervous system during limb regeneration.
This image was captured using a light sheet microscope.
Related to images 6928 and 6932.
This image was captured using a light sheet microscope.
Related to images 6928 and 6932.
Prayag Murawala, MDI Biological Laboratory and Hannover Medical School.
View Media
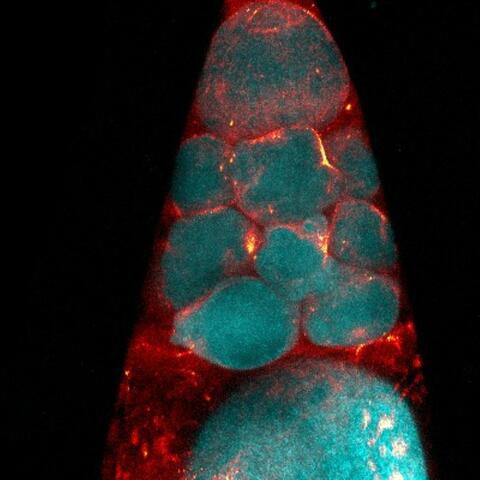
6753: Fruit fly nurse cells during egg development
6753: Fruit fly nurse cells during egg development
In many animals, the egg cell develops alongside sister cells. These sister cells are called nurse cells in the fruit fly (Drosophila melanogaster), and their job is to “nurse” an immature egg cell, or oocyte. Toward the end of oocyte development, the nurse cells transfer all their contents into the oocyte in a process called nurse cell dumping. This process involves significant shape changes on the part of the nurse cells (blue), which are powered by wavelike activity of the protein myosin (red). This image was captured using a confocal laser scanning microscope. Related to video 6754.
Adam C. Martin, Massachusetts Institute of Technology.
View Media
