Switch to List View
Image and Video Gallery
This is a searchable collection of scientific photos, illustrations, and videos. The images and videos in this gallery are licensed under Creative Commons Attribution Non-Commercial ShareAlike 3.0. This license lets you remix, tweak, and build upon this work non-commercially, as long as you credit and license your new creations under identical terms.
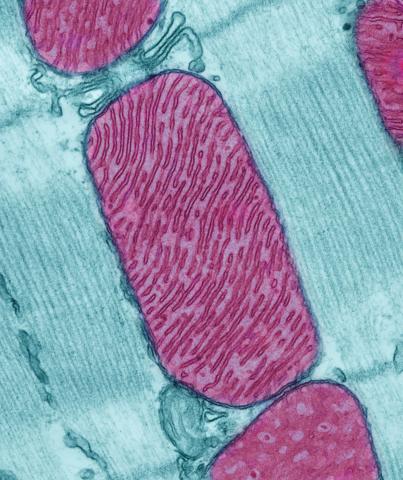
3661: Mitochondria from rat heart muscle cell
3661: Mitochondria from rat heart muscle cell
These mitochondria (red) are from the heart muscle cell of a rat. Mitochondria have an inner membrane that folds in many places (and that appears here as striations). This folding vastly increases the surface area for energy production. Nearly all our cells have mitochondria. Related to image 3664.
National Center for Microscopy and Imaging Research
View Media
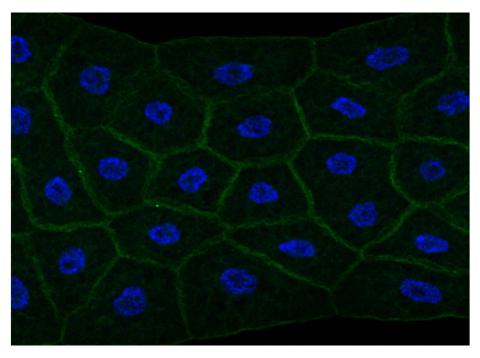
2757: Draper, shown in the fatbody of a Drosophila melanogaster larva
2757: Draper, shown in the fatbody of a Drosophila melanogaster larva
The fly fatbody is a nutrient storage and mobilization organ akin to the mammalian liver. The engulfment receptor Draper (green) is located at the cell surface of fatbody cells. The cell nuclei are shown in blue.
Christina McPhee and Eric Baehrecke, University of Massachusetts Medical School
View Media

3732: A molecular interaction network in yeast 2
3732: A molecular interaction network in yeast 2
The image visualizes a part of the yeast molecular interaction network. The lines in the network represent connections among genes (shown as little dots) and different-colored networks indicate subnetworks, for instance, those in specific locations or pathways in the cell. Researchers use gene or protein expression data to build these networks; the network shown here was visualized with a program called Cytoscape. By following changes in the architectures of these networks in response to altered environmental conditions, scientists can home in on those genes that become central "hubs" (highly connected genes), for example, when a cell encounters stress. They can then further investigate the precise role of these genes to uncover how a cell's molecular machinery deals with stress or other factors. Related to images 3730 and 3733.
Keiichiro Ono, UCSD
View Media
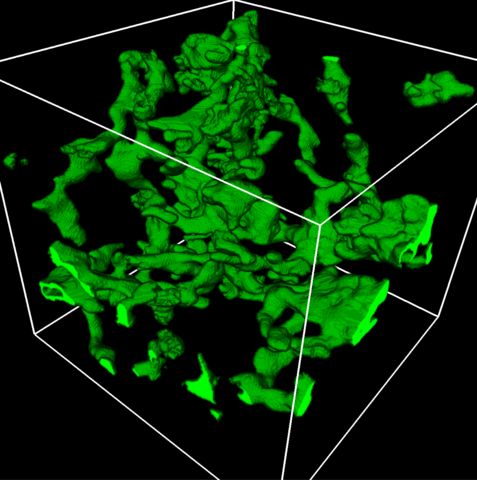
5857: 3D reconstruction of a tubular matrix in peripheral endoplasmic reticulum
5857: 3D reconstruction of a tubular matrix in peripheral endoplasmic reticulum
Detailed three-dimensional reconstruction of a tubular matrix in a thin section of the peripheral endoplasmic reticulum between the plasma membranes of the cell. The endoplasmic reticulum (ER) is a continuous membrane that extends like a net from the envelope of the nucleus outward to the cell membrane. The ER plays several roles within the cell, such as in protein and lipid synthesis and transport of materials between organelles. Shown here is a three-dimensional representation of the peripheral ER microtubules. Related to images 5855 and 5856
Jennifer Lippincott-Schwartz, Howard Hughes Medical Institute Janelia Research Campus, Virginia
View Media

1334: Aging book of life
1334: Aging book of life
Damage to each person's genome, often called the "Book of Life," accumulates with time. Such DNA mutations arise from errors in the DNA copying process, as well as from external sources, such as sunlight and cigarette smoke. DNA mutations are known to cause cancer and also may contribute to cellular aging.
Judith Stoffer
View Media
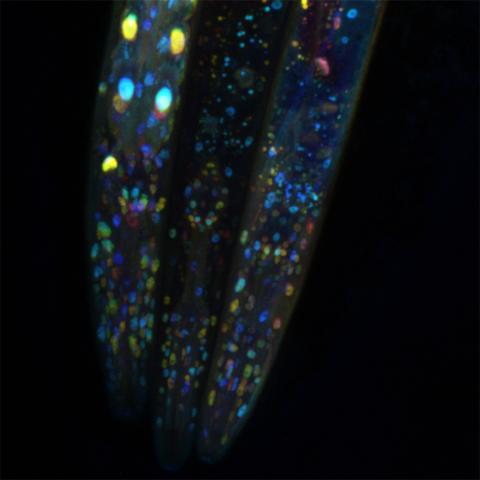
6532: Mosaicism in C. elegans (Black Background)
6532: Mosaicism in C. elegans (Black Background)
In the worm C. elegans, double-stranded RNA made in neurons can silence matching genes in a variety of cell types through the transport of RNA between cells. The head region of three worms that were genetically modified to express a fluorescent protein were imaged and the images were color-coded based on depth. The worm on the left lacks neuronal double-stranded RNA and thus every cell is fluorescent. In the middle worm, the expression of the fluorescent protein is silenced by neuronal double-stranded RNA and thus most cells are not fluorescent. The worm on the right lacks an enzyme that amplifies RNA for silencing. Surprisingly, the identities of the cells that depend on this enzyme for gene silencing are unpredictable. As a result, worms of identical genotype are nevertheless random mosaics for how the function of gene silencing is carried out. For more, see journal article and press release. Related to image 6534.
Snusha Ravikumar, Ph.D., University of Maryland, College Park, and Antony M. Jose, Ph.D., University of Maryland, College Park
View Media
1085: Natcher Building 05
1085: Natcher Building 05
NIGMS staff are located in the Natcher Building on the NIH campus.
Alisa Machalek, National Institute of General Medical Sciences
View Media

2417: Fly by night
2417: Fly by night
This fruit fly expresses green fluorescent protein (GFP) in the same pattern as the period gene, a gene that regulates circadian rhythm and is expressed in all sensory neurons on the surface of the fly.
Jay Hirsh, University of Virginia
View Media
2797: Anti-tumor drug ecteinascidin 743 (ET-743), structure without hydrogens 04
2797: Anti-tumor drug ecteinascidin 743 (ET-743), structure without hydrogens 04
Ecteinascidin 743 (ET-743, brand name Yondelis), was discovered and isolated from a sea squirt, Ecteinascidia turbinata, by NIGMS grantee Kenneth Rinehart at the University of Illinois. It was synthesized by NIGMS grantees E.J. Corey and later by Samuel Danishefsky. Multiple versions of this structure are available as entries 2790-2797.
Timothy Jamison, Massachusetts Institute of Technology
View Media
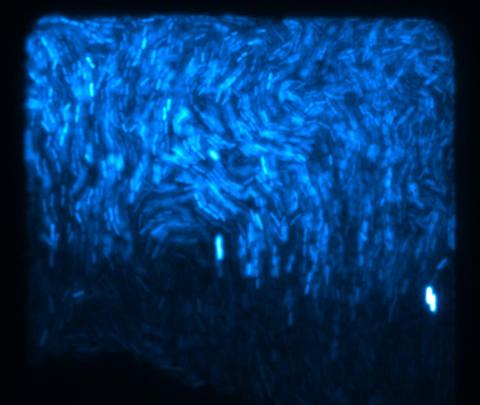
3268: Fluorescent E. coli bacteria
3268: Fluorescent E. coli bacteria
Bioengineers were able to coax bacteria to blink in unison on microfluidic chips. They called each blinking bacterial colony a biopixel. Thousands of fluorescent E. coli bacteria, shown here, make up a biopixel. Related to images 3265 and 3266. From a UC San Diego news release, "Researchers create living 'neon signs' composed of millions of glowing bacteria."
Jeff Hasty Lab, UC San Diego
View Media
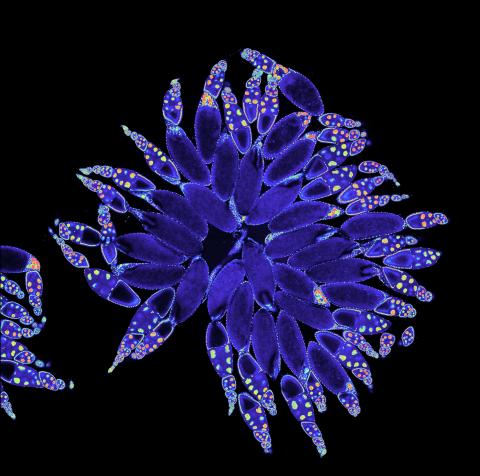
3607: Fruit fly ovary
3607: Fruit fly ovary
A fruit fly ovary, shown here, contains as many as 20 eggs. Fruit flies are not merely tiny insects that buzz around overripe fruit—they are a venerable scientific tool. Research on the flies has shed light on many aspects of human biology, including biological rhythms, learning, memory, and neurodegenerative diseases. Another reason fruit flies are so useful in a lab (and so successful in fruit bowls) is that they reproduce rapidly. About three generations can be studied in a single month.
Related to image 3656. This image was part of the Life: Magnified exhibit that ran from June 3, 2014, to January 21, 2015, at Dulles International Airport.
Related to image 3656. This image was part of the Life: Magnified exhibit that ran from June 3, 2014, to January 21, 2015, at Dulles International Airport.
Denise Montell, Johns Hopkins University and University of California, Santa Barbara
View Media
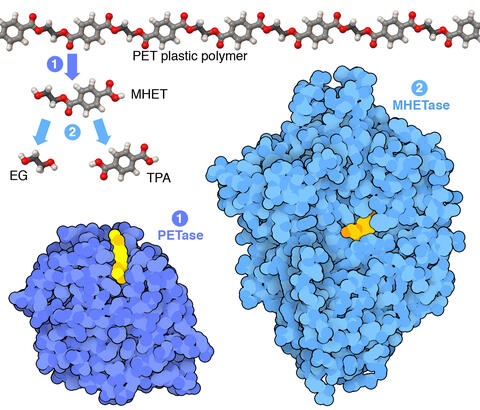
7000: Plastic-eating enzymes
7000: Plastic-eating enzymes
PETase enzyme degrades polyester plastic (polyethylene terephthalate, or PET) into monohydroxyethyl terephthalate (MHET). Then, MHETase enzyme degrades MHET into its constituents ethylene glycol (EG) and terephthalic acid (TPA).
Find these in the RCSB Protein Data Bank: PET hydrolase (PDB entry 5XH3) and MHETase (PDB entry 6QGA).
Find these in the RCSB Protein Data Bank: PET hydrolase (PDB entry 5XH3) and MHETase (PDB entry 6QGA).
Amy Wu and Christine Zardecki, RCSB Protein Data Bank.
View Media
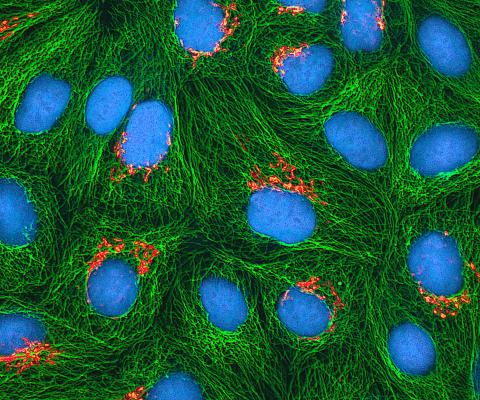
3522: HeLa cells
3522: HeLa cells
Multiphoton fluorescence image of cultured HeLa cells with a fluorescent protein targeted to the Golgi apparatus (orange), microtubules (green) and counterstained for DNA (cyan). Nikon RTS2000MP custom laser scanning microscope. See related images 3518, 3519, 3520, 3521.
National Center for Microscopy and Imaging Research (NCMIR)
View Media
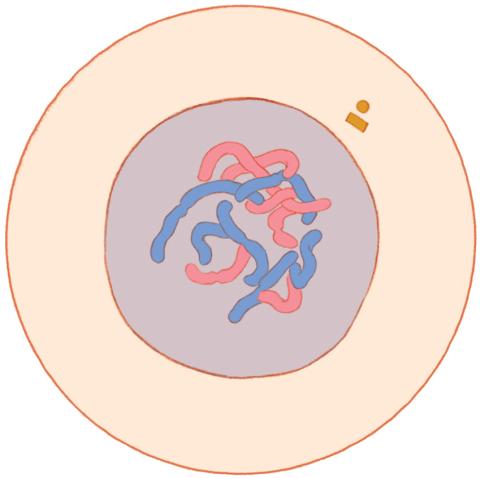
1316: Mitosis - interphase
1316: Mitosis - interphase
A cell in interphase, at the start of mitosis: Chromosomes duplicate, and the copies remain attached to each other. Mitosis is responsible for growth and development, as well as for replacing injured or worn out cells throughout the body. For simplicity, mitosis is illustrated here with only six chromosomes.
Judith Stoffer
View Media
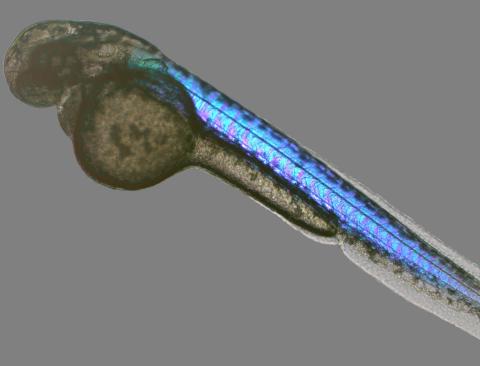
6897: Zebrafish embryo
6897: Zebrafish embryo
A zebrafish embryo showing its natural colors. Zebrafish have see-through eggs and embryos, making them ideal research organisms for studying the earliest stages of development. This image was taken in transmitted light under a polychromatic polarizing microscope.
Michael Shribak, Marine Biological Laboratory/University of Chicago.
View Media
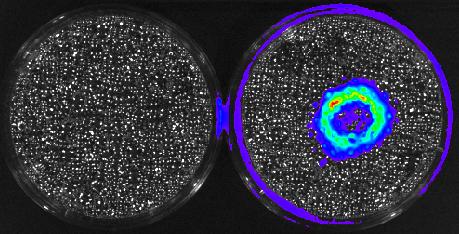
3480: Cancer Cells Glowing from Luciferin
3480: Cancer Cells Glowing from Luciferin
The activator cancer cell culture, right, contains a chemical that causes the cells to emit light when in the presence of immune cells.
Mark Sellmyer, Stanford University School of Medicine
View Media
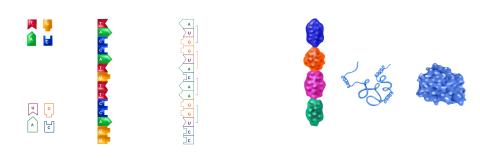
2509: From DNA to Protein
2509: From DNA to Protein
Nucleotides in DNA are copied into RNA, where they are read three at a time to encode the amino acids in a protein. Many parts of a protein fold as the amino acids are strung together.
See image 2510 for a labeled version of this illustration.
Featured in The Structures of Life.
See image 2510 for a labeled version of this illustration.
Featured in The Structures of Life.
Crabtree + Company
View Media
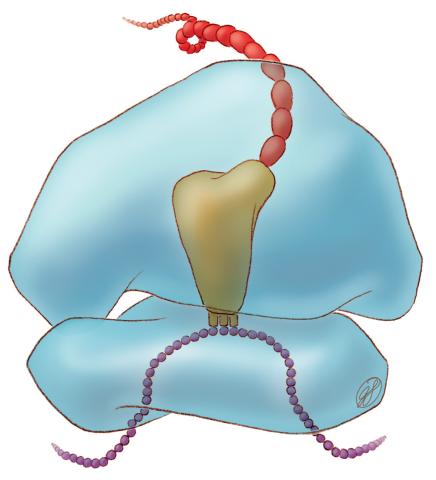
1281: Translation
1281: Translation
Ribosomes manufacture proteins based on mRNA instructions. Each ribosome reads mRNA, recruits tRNA molecules to fetch amino acids, and assembles the amino acids in the proper order.
Judith Stoffer
View Media
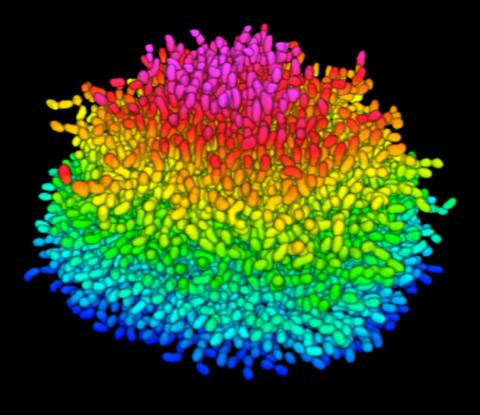
5825: A Growing Bacterial Biofilm
5825: A Growing Bacterial Biofilm
A growing Vibrio cholerae (cholera) biofilm. Cholera bacteria form colonies called biofilms that enable them to resist antibiotic therapy within the body and other challenges to their growth.
Each slightly curved comma shape represents an individual bacterium from assembled confocal microscopy images. Different colors show each bacterium’s position in the biofilm in relation to the surface on which the film is growing.
Each slightly curved comma shape represents an individual bacterium from assembled confocal microscopy images. Different colors show each bacterium’s position in the biofilm in relation to the surface on which the film is growing.
Jing Yan, Ph.D., and Bonnie Bassler, Ph.D., Department of Molecular Biology, Princeton University, Princeton, NJ.
View Media
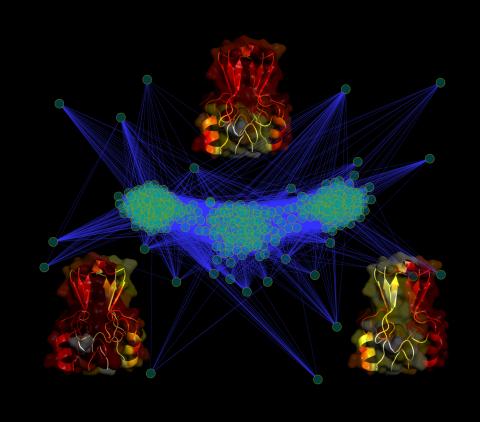
3295: Cluster analysis of mysterious protein
3295: Cluster analysis of mysterious protein
Researchers use cluster analysis to study protein shape and function. Each green circle represents one potential shape of the protein mitoNEET. The longer the blue line between two circles, the greater the differences between the shapes. Most shapes are similar; they fall into three clusters that are represented by the three images of the protein. From a Rice University news release. Graduate student Elizabeth Baxter and Patricia Jennings, professor of chemistry and biochemistry at UCSD, collaborated with José Onuchic, a physicist at Rice University, on this work.
Patricia Jennings and Elizabeth Baxter, University of California, San Diego
View Media
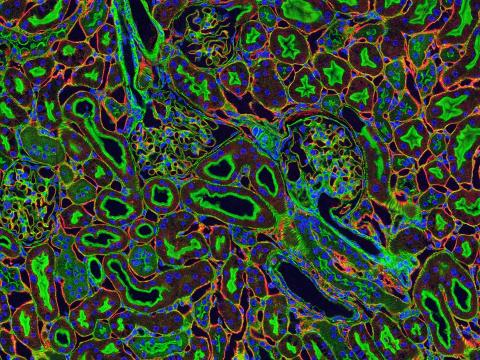
3675: NCMIR kidney-1
3675: NCMIR kidney-1
Stained kidney tissue. The kidney is an essential organ responsible for disposing wastes from the body and for maintaining healthy ion levels in the blood. It also secretes two hormones, erythropoietin (EPO) and calcitriol (a derivative of vitamin D), into the blood. It works like a purifier by pulling break-down products of metabolism, such as urea and ammonium, from the blood stream for excretion in urine. Related to image 3725.
Tom Deerinck, National Center for Microscopy and Imaging Research (NCMIR)
View Media
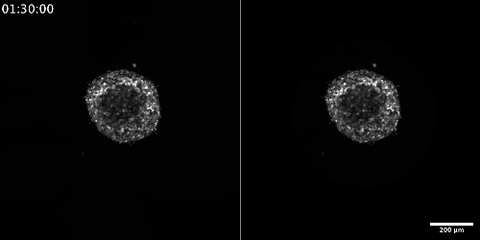
6986: Breast cancer cells change migration phenotypes
6986: Breast cancer cells change migration phenotypes
Cancer cells can change their migration phenotype, which includes their shape and the way that they move to invade different tissues. This movie shows breast cancer cells forming a tumor spheroid—a 3D ball of cancer cells—and invading the surrounding tissue. Images were taken using a laser scanning confocal microscope, and artificial intelligence (AI) models were used to segment and classify the images by migration phenotype. On the right side of the video, each phenotype is represented by a different color, as recognized by the AI program based on identifiable characteristics of those phenotypes. The movie demonstrates how cancer cells can use different migration modes during growth and metastasis—the spreading of cancer cells within the body.
Bo Sun, Oregon State University.
View Media

2362: Automated crystal screening system
2362: Automated crystal screening system
Automated crystal screening systems such as the one shown here are becoming a common feature at synchrotron and other facilities where high-throughput crystal structure determination is being carried out. These systems rapidly screen samples to identify the best candidates for further study.
Southeast Collaboratory for Structural Genomics
View Media

3689: Computer sketch of bird-and-flower DNA origami
3689: Computer sketch of bird-and-flower DNA origami
A computer-generated sketch of a DNA origami folded into a flower-and-bird structure. See also related image 3690.
Hao Yan, Arizona State University
View Media
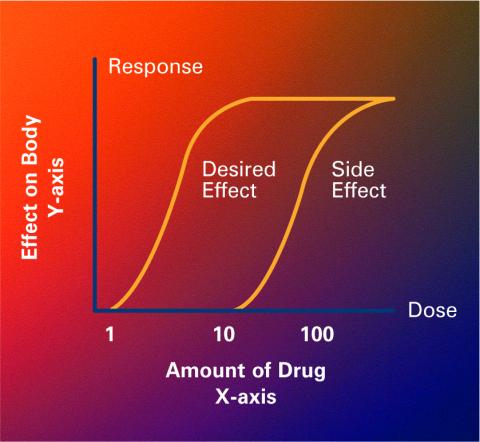
2533: Dose response curves
2533: Dose response curves
Dose-response curves determine how much of a drug (X-axis) causes a particular effect, or a side effect, in the body (Y-axis). Featured in Medicines By Design.
Crabtree + Company
View Media
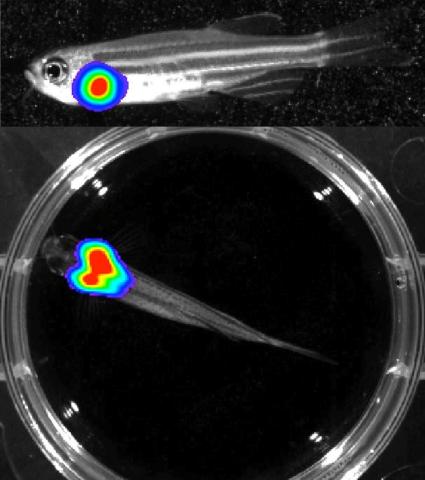
3556: Bioluminescent imaging in adult zebrafish - lateral and overhead view
3556: Bioluminescent imaging in adult zebrafish - lateral and overhead view
Luciferase-based imaging enables visualization and quantification of internal organs and transplanted cells in live adult zebrafish. In this image, a cardiac muscle-restricted promoter drives firefly luciferase expression. This is the lateral and overhead (Bottom) view.
For imagery of the overhead view go to 3557.
For imagery of the lateral view go to 3558.
For more information about the illumated area go to 3559.
For imagery of the overhead view go to 3557.
For imagery of the lateral view go to 3558.
For more information about the illumated area go to 3559.
Kenneth Poss, Duke University
View Media
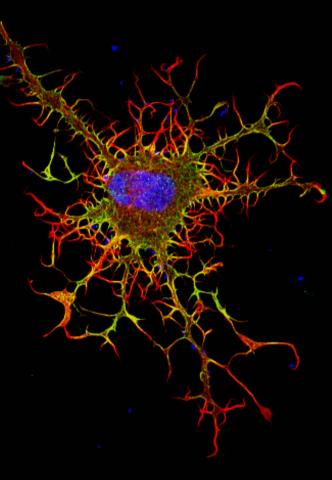
3613: Abnormal, spiky fibroblast
3613: Abnormal, spiky fibroblast
This is a fibroblast, a connective tissue cell that plays an important role in wound healing. Normal fibroblasts have smooth edges. In contrast, this spiky cell is missing a protein that is necessary for proper construction of the cell's skeleton. Its jagged shape makes it impossible for the cell to move normally. In addition to compromising wound healing, abnormal cell movement can lead to birth defects, faulty immune function, and other health problems.
This image was part of the Life: Magnified exhibit that ran from June 3, 2014, to January 21, 2015, at Dulles International Airport.
This image was part of the Life: Magnified exhibit that ran from June 3, 2014, to January 21, 2015, at Dulles International Airport.
Praveen Suraneni, Stowers Institute for Medical Research, Kansas City, Mo.
View Media
1056: Skin cross-section
1056: Skin cross-section
Cross-section of skin anatomy shows layers and different tissue types.
National Institutes of Health Medical Arts
View Media

2576: Cone snail shell
2576: Cone snail shell
A shell from the venomous cone snail Conus omaria, which lives in the Pacific and Indian oceans and eats other snails. University of Utah scientists discovered a new toxin in this snail species' venom, and say it will be a useful tool in designing new medicines for a variety of brain disorders, including Alzheimer's and Parkinson's diseases, depression, nicotine addiction and perhaps schizophrenia.
Kerry Matz, University of Utah
View Media
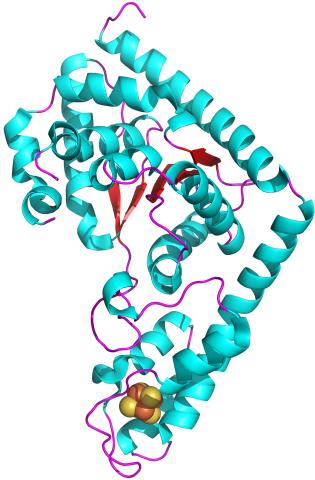
2483: Trp_RS - tryptophanyl tRNA-synthetase family of enzymes
2483: Trp_RS - tryptophanyl tRNA-synthetase family of enzymes
This image represents the structure of TrpRS, a novel member of the tryptophanyl tRNA-synthetase family of enzymes. By helping to link the amino acid tryptophan to a tRNA molecule, TrpRS primes the amino acid for use in protein synthesis. A cluster of iron and sulfur atoms (orange and red spheres) was unexpectedly found in the anti-codon domain, a key part of the molecule, and appears to be critical for the function of the enzyme. TrpRS was discovered in Thermotoga maritima, a rod-shaped bacterium that flourishes in high temperatures.
View Media
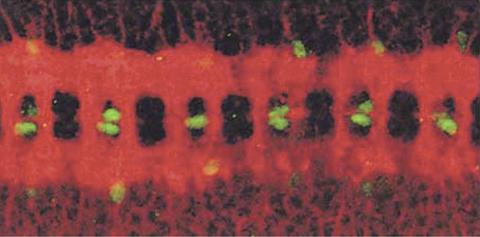
1091: Nerve and glial cells in fruit fly embryo
1091: Nerve and glial cells in fruit fly embryo
Glial cells (stained green) in a fruit fly developing embryo have survived thanks to a signaling pathway initiated by neighboring nerve cells (stained red).
Hermann Steller, Rockefeller University
View Media
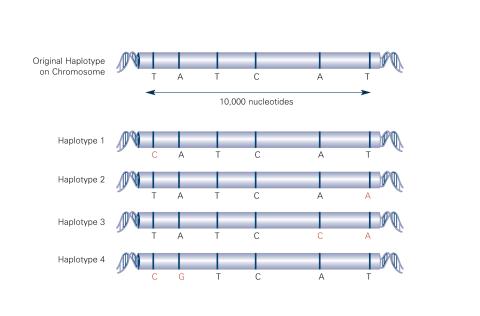
2567: Haplotypes (with labels)
2567: Haplotypes (with labels)
Haplotypes are combinations of gene variants that are likely to be inherited together within the same chromosomal region. In this example, an original haplotype (top) evolved over time to create three newer haplotypes that each differ by a few nucleotides (red). See image 2566 for an unlabeled version of this illustration. Featured in The New Genetics.
Crabtree + Company
View Media
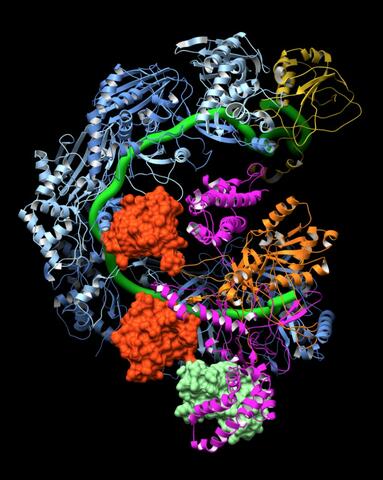
6352: CRISPR surveillance complex
6352: CRISPR surveillance complex
This image shows how the CRISPR surveillance complex is disabled by two copies of anti-CRISPR protein AcrF1 (red) and one AcrF2 (light green). These anti-CRISPRs block access to the CRISPR RNA (green tube) preventing the surveillance complex from scanning and targeting invading viral DNA for destruction.
NRAMM National Resource for Automated Molecular Microscopy http://nramm.nysbc.org/nramm-images/ Source: Bridget Carragher
View Media
3266: Biopixels
3266: Biopixels
Bioengineers were able to coax bacteria to blink in unison on microfluidic chips. This image shows a small chip with about 500 blinking bacterial colonies or biopixels. Related to images 3265 and 3268. From a UC San Diego news release, "Researchers create living 'neon signs' composed of millions of glowing bacteria."
Jeff Hasty Lab, UC San Diego
View Media
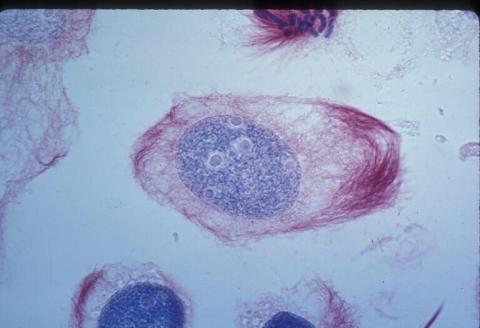
1012: Lily mitosis 02
1012: Lily mitosis 02
A light microscope image of a cell from the endosperm of an African globe lily (Scadoxus katherinae). This is one frame of a time-lapse sequence that shows cell division in action. The lily is considered a good organism for studying cell division because its chromosomes are much thicker and easier to see than human ones. Staining shows microtubules in red and chromosomes in blue.
Related to images 1010, 1011, 1013, 1014, 1015, 1016, 1017, 1018, 1019, and 1021.
Related to images 1010, 1011, 1013, 1014, 1015, 1016, 1017, 1018, 1019, and 1021.
Andrew S. Bajer, University of Oregon, Eugene
View Media
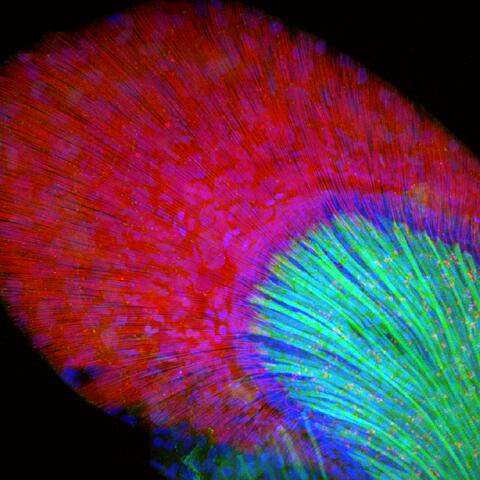
3598: Developing zebrafish fin
3598: Developing zebrafish fin
Originally from the waters of India, Nepal, and neighboring countries, zebrafish can now be found swimming in science labs (and home aquariums) throughout the world. This fish is a favorite study subject for scientists interested in how genes guide the early stages of prenatal development (including the developing fin shown here) and in the effects of environmental contamination on embryos.
In this image, green fluorescent protein (GFP) is expressed where the gene sox9b is expressed. Collagen (red) marks the fin rays, and DNA, stained with a dye called DAPI, is in blue. sox9b plays many important roles during development, including the building of the heart and brain, and is also necessary for skeletal development. At the University of Wisconsin, researchers have found that exposure to contaminants that bind the aryl-hydrocarbon receptor results in the downregulation of sox9b. Loss of sox9b severely disrupts development in zebrafish and causes a life-threatening disorder called campomelic dysplasia (CD) in humans. CD is characterized by cardiovascular, neural, and skeletal defects. By studying the roles of genes such as sox9b in zebrafish, scientists hope to better understand normal development in humans as well as how to treat developmental disorders and diseases.
This image was part of the Life: Magnified exhibit that ran from June 3, 2014, to January 21, 2015, at Dulles International Airport.
In this image, green fluorescent protein (GFP) is expressed where the gene sox9b is expressed. Collagen (red) marks the fin rays, and DNA, stained with a dye called DAPI, is in blue. sox9b plays many important roles during development, including the building of the heart and brain, and is also necessary for skeletal development. At the University of Wisconsin, researchers have found that exposure to contaminants that bind the aryl-hydrocarbon receptor results in the downregulation of sox9b. Loss of sox9b severely disrupts development in zebrafish and causes a life-threatening disorder called campomelic dysplasia (CD) in humans. CD is characterized by cardiovascular, neural, and skeletal defects. By studying the roles of genes such as sox9b in zebrafish, scientists hope to better understand normal development in humans as well as how to treat developmental disorders and diseases.
This image was part of the Life: Magnified exhibit that ran from June 3, 2014, to January 21, 2015, at Dulles International Airport.
Jessica Plavicki
View Media
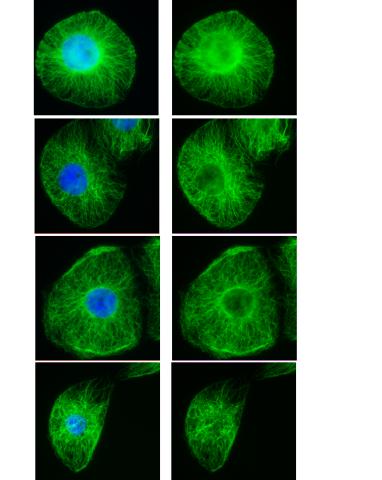
3443: Interphase in Xenopus frog cells
3443: Interphase in Xenopus frog cells
These images show frog cells in interphase. The cells are Xenopus XL177 cells, which are derived from tadpole epithelial cells. The microtubules are green and the chromosomes are blue. Related to 3442.
Claire Walczak, who took them while working as a postdoc in the laboratory of Timothy Mitchison.
View Media
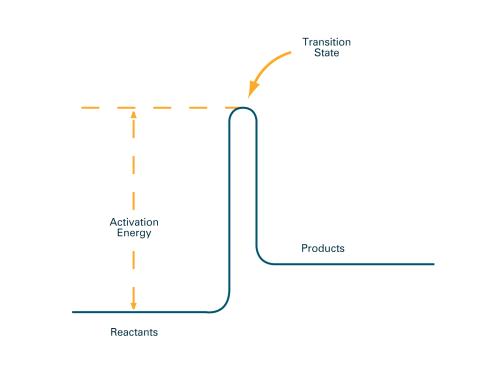
2526: Activation energy (with labels)
2526: Activation energy (with labels)
To become products, reactants must overcome an energy hill. See image 2525 for an unlabeled version of this illustration. Featured in The Chemistry of Health.
Crabtree + Company
View Media
