Switch to List View
Image and Video Gallery
This is a searchable collection of scientific photos, illustrations, and videos. The images and videos in this gallery are licensed under Creative Commons Attribution Non-Commercial ShareAlike 3.0. This license lets you remix, tweak, and build upon this work non-commercially, as long as you credit and license your new creations under identical terms.
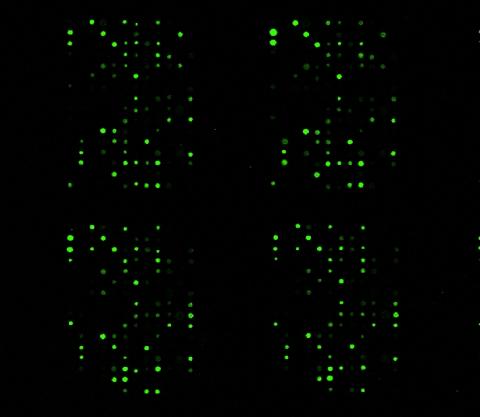
1265: Glycan arrays
1265: Glycan arrays
The signal is obtained by allowing proteins in human serum to interact with glycan (polysaccharide) arrays. The arrays are shown in replicate so the pattern is clear. Each spot contains a specific type of glycan. Proteins have bound to the spots highlighted in green.
Ola Blixt, Scripps Research Institute
View Media
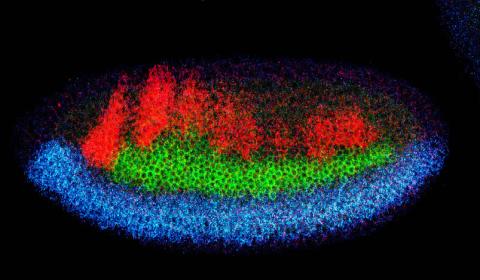
2327: Neural development
2327: Neural development
Using techniques that took 4 years to design, a team of developmental biologists showed that certain proteins can direct the subdivision of fruit fly and chicken nervous system tissue into the regions depicted here in blue, green, and red. Molecules called bone morphogenetic proteins (BMPs) helped form this fruit fly embryo. While scientists knew that BMPs play a major role earlier in embryonic development, they didn't know how the proteins help organize nervous tissue. The findings suggest that BMPs are part of an evolutionarily conserved mechanism for organizing the nervous system. The National Institute of Neurological Disorders and Stroke also supported this work.
Mieko Mizutani and Ethan Bier, University of California, San Diego, and Henk Roelink, University of Washington
View Media
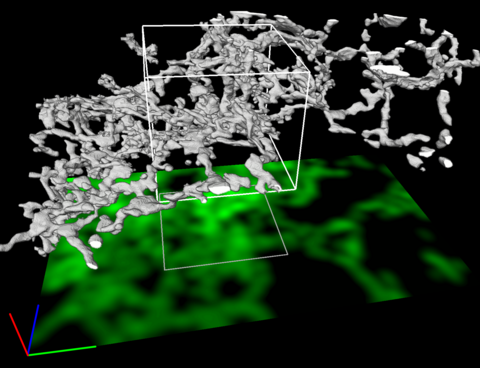
5856: Dense tubular matrices in the peripheral endoplasmic reticulum (ER) 2
5856: Dense tubular matrices in the peripheral endoplasmic reticulum (ER) 2
Three-dimensional reconstruction of a tubular matrix in a thin section of the peripheral endoplasmic reticulum between the plasma membranes of the cell. The endoplasmic reticulum (ER) is a continuous membrane that extends like a net from the envelope of the nucleus outward to the cell membrane. The ER plays several roles within the cell, such as in protein and lipid synthesis and transport of materials between organelles. Shown here are super-resolution microscopic images of the peripheral ER showing the structure of an ER tubular matrix between the plasma membranes of the cell. See image 5857 for a more detailed view of the area outlined in white in this image. For another view of the ER tubular matrix see image 5855
Jennifer Lippincott-Schwartz, Howard Hughes Medical Institute Janelia Research Campus, Virginia
View Media

2709: Retroviruses as fossils
2709: Retroviruses as fossils
DNA doesn't leave a fossil record in stone, the way bones do. Instead, the DNA code itself holds the best evidence for organisms' genetic history. Some of the most telling evidence about genetic history comes from retroviruses, the remnants of ancient viral infections.
Emily Harrington, science illustrator
View Media
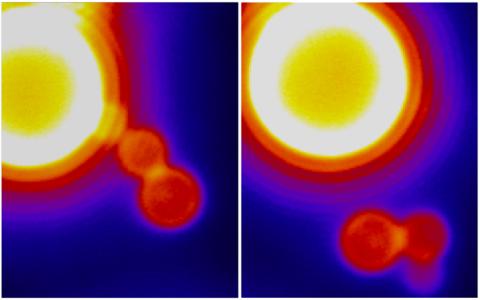
3448: Dynamin Fission
3448: Dynamin Fission
Time lapse series shows short dynamin assemblies (not visible) constricting a lipid tube to make a "beads on a string" appearance, then cutting off one of the beads i.e., catalyzing membrane fission). The lipids are fluorescent (artificially colored). Ramachandran R, Pucadyil T.J., Liu Y.W., Acharya S., Leonard M., Lukiyanchuk V., Schmid S.L. 2009. Membrane insertion of the pleckstrin homology domain variable loop 1 is critical for dynamin-catalyzed vesicle scission. Mol Biol Cell. 2009 20:4630-9.
Ramachandran, Pucadyil et al. , The Scripps Research Institute
View Media
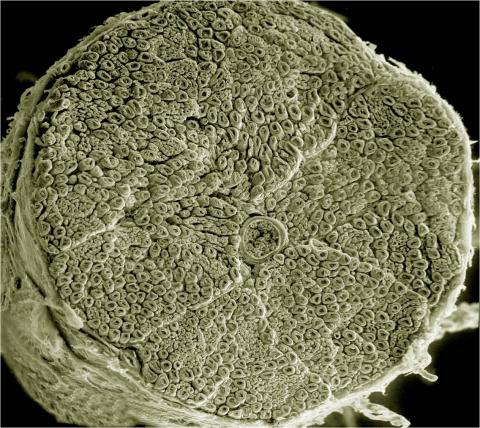
3387: NCMIR human spinal nerve
3387: NCMIR human spinal nerve
Spinal nerves are part of the peripheral nervous system. They run within the spinal column to carry nerve signals to and from all parts of the body. The spinal nerves enable all the movements we do, from turning our heads to wiggling our toes, control the movements of our internal organs, such as the colon and the bladder, as well as allow us to feel touch and the location of our limbs.
Tom Deerinck, National Center for Microscopy and Imaging Research (NCMIR)
View Media
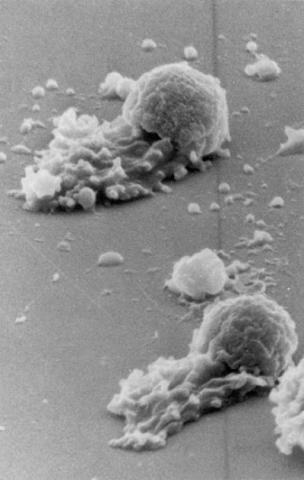
3489: Worm sperm
3489: Worm sperm
To develop a system for studying cell motility in unnatrual conditions -- a microscope slide instead of the body -- Tom Roberts and Katsuya Shimabukuro at Florida State University disassembled and reconstituted the motility parts used by worm sperm cells.
Tom Roberts, Florida State University
View Media
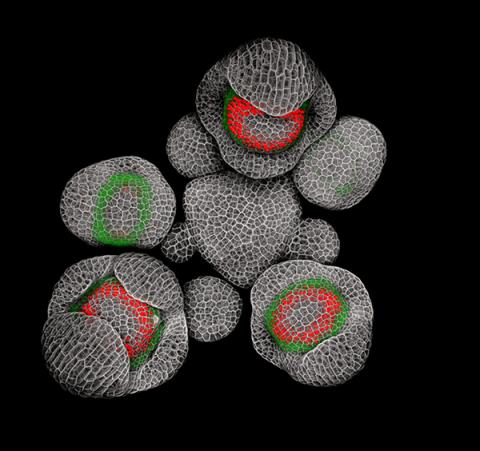
3743: Developing Arabidopsis flower buds
3743: Developing Arabidopsis flower buds
Flower development is a carefully orchestrated, genetically programmed process that ensures that the male (stamen) and female (pistil) organs form in the right place and at the right time in the flower. In this image of young Arabidopsis flower buds, the gene SUPERMAN (red) is activated at the boundary between the cells destined to form the male and female parts. SUPERMAN activity prevents the central cells, which will ultimately become the female pistil, from activating the gene APETALA3 (green), which induces formation of male flower organs. The goal of this research is to find out how plants maintain cells (called stem cells) that have the potential to develop into any type of cell and how genetic and environmental factors cause stem cells to develop and specialize into different cell types. This work informs future studies in agriculture, medicine and other fields.
Nathanaël Prunet, Caltech
View Media
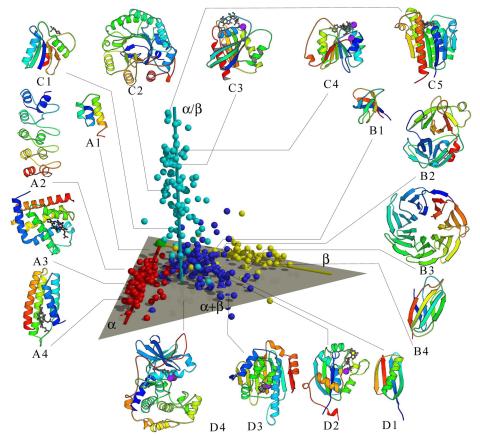
2367: Map of protein structures 02
2367: Map of protein structures 02
A global "map of the protein structure universe" indicating the positions of specific proteins. The preponderance of small, less-structured proteins near the origin, with the more highly structured, large proteins towards the ends of the axes, may suggest the evolution of protein structures.
Berkeley Structural Genomics Center, PSI
View Media

2702: Thermotoga maritima and its metabolic network
2702: Thermotoga maritima and its metabolic network
A combination of protein structures determined experimentally and computationally shows us the complete metabolic network of a heat-loving bacterium.
View Media
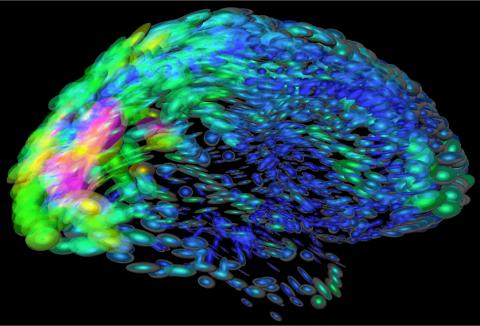
2419: Mapping brain differences
2419: Mapping brain differences
This image of the human brain uses colors and shapes to show neurological differences between two people. The blurred front portion of the brain, associated with complex thought, varies most between the individuals. The blue ovals mark areas of basic function that vary relatively little. Visualizations like this one are part of a project to map complex and dynamic information about the human brain, including genes, enzymes, disease states, and anatomy. The brain maps represent collaborations between neuroscientists and experts in math, statistics, computer science, bioinformatics, imaging, and nanotechnology.
Arthur Toga, University of California, Los Angeles
View Media

2414: Pig trypsin (3)
2414: Pig trypsin (3)
Crystals of porcine trypsin protein created for X-ray crystallography, which can reveal detailed, three-dimensional protein structures.
Alex McPherson, University of California, Irvine
View Media
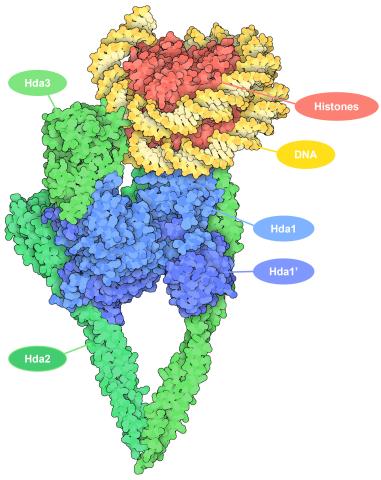
7001: Histone deacetylases
7001: Histone deacetylases
The human genome contains much of the information needed for every cell in the body to function. However, different types of cells often need different types of information. Access to DNA is controlled, in part, by how tightly it’s wrapped around proteins called histones to form nucleosomes. The complex shown here, from yeast cells (PDB entry 6Z6P), includes several histone deacetylase (HDAC) enzymes (green and blue) bound to a nucleosome (histone proteins in red; DNA in yellow). The yeast HDAC enzymes are similar to the human enzymes. Two enzymes form a V-shaped clamp (green) that holds the other others, a dimer of the Hda1 enzymes (blue). In this assembly, Hda1 is activated and positioned to remove acetyl groups from histone tails.
Amy Wu and Christine Zardecki, RCSB Protein Data Bank.
View Media
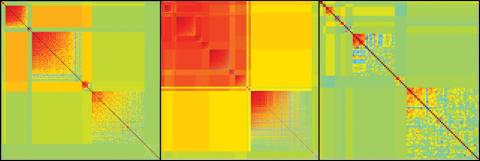
2588: Genetic patchworks
2588: Genetic patchworks
Each point in these colorful patchworks represents the correlation between two sleep-associated genes in fruit flies. Vibrant reds and oranges represent high and intermediate degrees of association between the genes, respectively. Genes in these areas show similar activity patterns in different fly lines. Cool blues represent gene pairs where one partner's activity is high and the other's is low. The green areas show pairs with activities that are not correlated. These quilt-like depictions help illustrate a recent finding that genes act in teams to influence sleep patterns.
Susan Harbison and Trudy Mackay, North Carolina State University
View Media
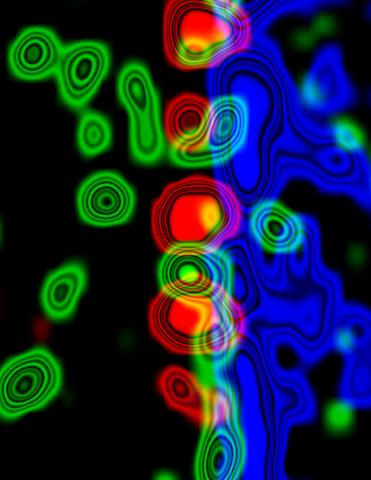
3734: Molecular interactions at the astrocyte nuclear membrane
3734: Molecular interactions at the astrocyte nuclear membrane
These ripples of color represent the outer membrane of the nucleus inside an astrocyte, a star-shaped cell inside the brain. Some proteins (green) act as keys to unlock other proteins (red) that form gates to let small molecules in and out of the nucleus (blue). Visualizing these different cell components at the boundary of the astrocyte nucleus enables researchers to study the molecular and physiological basis of neurological disorders, such as hydrocephalus, a condition in which too much fluid accumulates in the brain, and scar formation in brain tissue leading to abnormal neuronal activity affecting learning and memory. Scientists have now identified a pathway may be common to many of these brain diseases and begun to further examine it to find ways to treat certain brain diseases and injuries.
Katerina Akassoglou, Gladstone Institute for Neurological Disease & UCSF
View Media
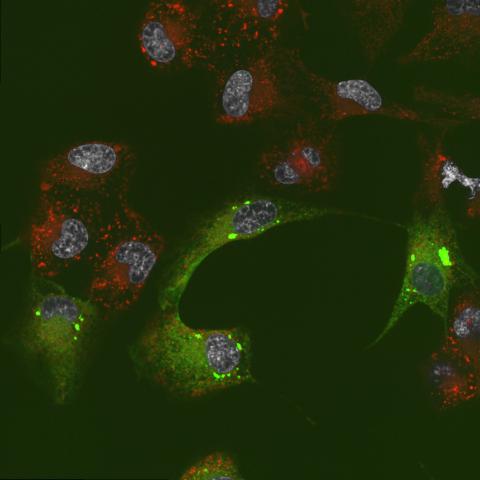
6774: Endoplasmic reticulum abnormalities 2
6774: Endoplasmic reticulum abnormalities 2
Human cells with the gene that codes for the protein FIT2 deleted. After an experimental intervention, they are expressing a nonfunctional version of FIT2, shown in green. The lack of functional FIT2 affected the structure of the endoplasmic reticulum (ER), and the nonfunctional protein clustered in ER membrane aggregates, seen as large bright-green spots. Lipid droplets are shown in red, and the nucleus is visible in gray. This image was captured using a confocal microscope. Related to image 6773.
Michel Becuwe, Harvard University.
View Media
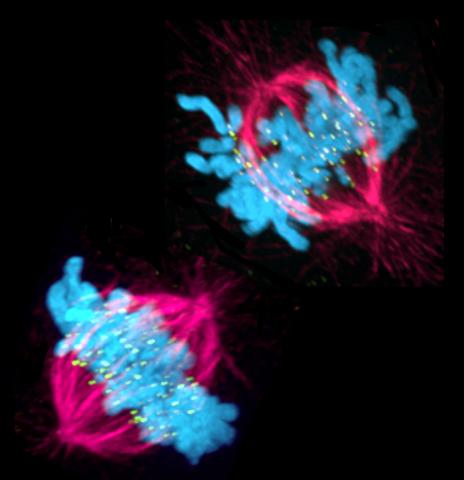
3541: Cell in two stages of division
3541: Cell in two stages of division
This image shows a cell in two stages of division: prometaphase (top) and metaphase (bottom). To form identical daughter cells, chromosome pairs (blue) separate via the attachment of microtubules made up of tubulin proteins (pink) to specialized structures on centromeres (green).
Lilian Kabeche, Dartmouth
View Media
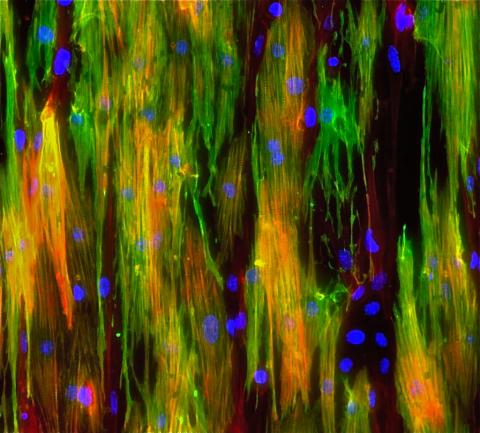
3283: Mouse heart muscle cells 02
3283: Mouse heart muscle cells 02
This image shows neonatal mouse heart cells. These cells were grown in the lab on a chip that aligns the cells in a way that mimics what is normally seen in the body. Green shows the muscle protein toponin I. Red indicates the muscle protein actin, and blue indicates the cell nuclei. The work shown here was part of a study attempting to grow heart tissue in the lab to repair damage after a heart attack. Image and caption information courtesy of the California Institute for Regenerative Medicine. Related to images 3281 and 3282.
Kara McCloskey lab, University of California, Merced, via CIRM
View Media
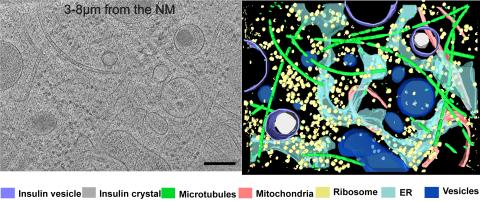
6608: Cryo-ET cross-section of a rat pancreas cell
6608: Cryo-ET cross-section of a rat pancreas cell
On the left, a cross-section slice of a rat pancreas cell captured using cryo-electron tomography (cryo-ET). On the right, a 3D, color-coded version of the image highlighting cell structures. Visible features include microtubules (neon-green rods), ribosomes (small yellow circles), and vesicles (dark-blue circles). These features are surrounded by the partially visible endoplasmic reticulum (light blue). The black line at the bottom right of the left image represents 200 nm. Related to image 6607.
Xianjun Zhang, University of Southern California.
View Media
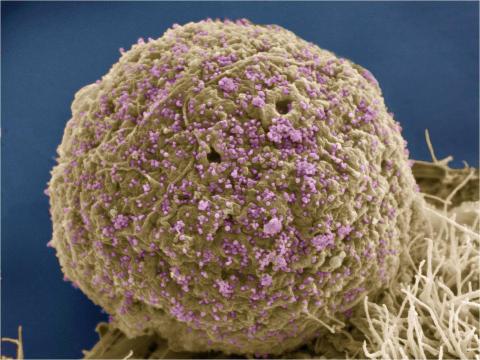
3386: HIV Infected Cell
3386: HIV Infected Cell
The human immunodeficiency virus (HIV), shown here as tiny purple spheres, causes the disease known as AIDS (for acquired immunodeficiency syndrome). HIV can infect multiple cells in your body, including brain cells, but its main target is a cell in the immune system called the CD4 lymphocyte (also called a T-cell or CD4 cell).
Tom Deerinck, National Center for Microscopy and Imaging Research (NCMIR)
View Media
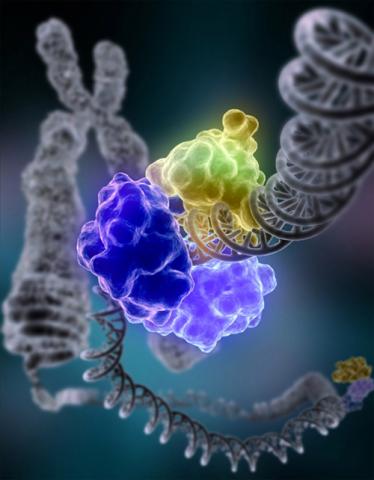
2330: Repairing DNA
2330: Repairing DNA
Like a watch wrapped around a wrist, a special enzyme encircles the double helix to repair a broken strand of DNA. Without molecules that can mend such breaks, cells can malfunction, die, or become cancerous. Related to image 3493.
Tom Ellenberger, Washington University School of Medicine
View Media
2432: ARTS triggers apoptosis
2432: ARTS triggers apoptosis
Cell showing overproduction of the ARTS protein (red). ARTS triggers apoptosis, as shown by the activation of caspase-3 (green) a key tool in the cell's destruction. The nucleus is shown in blue. Image is featured in October 2015 Biomedical Beat blog post Cool Images: A Halloween-Inspired Cell Collection.
Hermann Steller, Rockefeller University
View Media

7011: Hawaiian bobtail squid
7011: Hawaiian bobtail squid
An adult Hawaiian bobtail squid, Euprymna scolopes, swimming next to a submerged hand.
Related to image 7010 and video 7012.
Related to image 7010 and video 7012.
Margaret J. McFall-Ngai, Carnegie Institution for Science/California Institute of Technology, and Edward G. Ruby, California Institute of Technology.
View Media
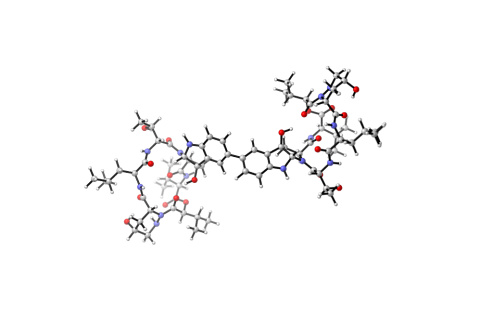
6848: Himastatin
6848: Himastatin
A model of the molecule himastatin, which was first isolated from the bacterium Streptomyces himastatinicus. Himastatin shows antibiotic activity. The researchers who created this image developed a new, more concise way to synthesize himastatin so it can be studied more easily.
More information about the research that produced this image can be found in the Science paper “Total synthesis of himastatin” by D’Angelo et al.
Related to image 6850 and video 6851.
More information about the research that produced this image can be found in the Science paper “Total synthesis of himastatin” by D’Angelo et al.
Related to image 6850 and video 6851.
Mohammad Movassaghi, Massachusetts Institute of Technology.
View Media

2683: GFP sperm
2683: GFP sperm
Fruit fly sperm cells glow bright green when they express the gene for green fluorescent protein (GFP).
View Media
2740: Early life of a protein
2740: Early life of a protein
This illustration represents the early life of a protein—specifically, apomyoglobin—as it is synthesized by a ribosome and emerges from the ribosomal tunnel, which contains the newly formed protein's conformation. The synthesis occurs in the complex swirl of the cell medium, filled with interactions among many molecules. Researchers in Silvia Cavagnero's laboratory are studying the structure and dynamics of newly made proteins and polypeptides using spectroscopic and biochemical techniques.
Silvia Cavagnero, University of Wisconsin, Madison
View Media
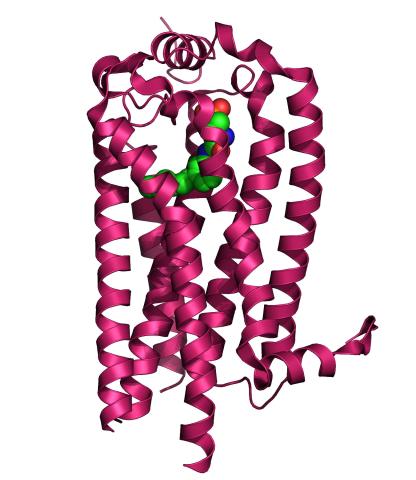
3362: Sphingolipid S1P1 receptor
3362: Sphingolipid S1P1 receptor
The receptor is shown bound to an antagonist, ML056.
Raymond Stevens, The Scripps Research Institute
View Media
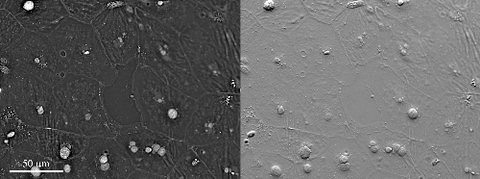
6899: Epithelial cell migration
6899: Epithelial cell migration
High-resolution time lapse of epithelial (skin) cell migration and wound healing. It shows an image taken every 13 seconds over the course of almost 14 minutes. The images were captured with quantitative orientation-independent differential interference contrast (DIC) microscope (left) and a conventional DIC microscope (right).
More information about the research that produced this video can be found in the Journal of Microscopy paper “An Orientation-Independent DIC Microscope Allows High Resolution Imaging of Epithelial Cell Migration and Wound Healing in a Cnidarian Model” by Malamy and Shribak.
More information about the research that produced this video can be found in the Journal of Microscopy paper “An Orientation-Independent DIC Microscope Allows High Resolution Imaging of Epithelial Cell Migration and Wound Healing in a Cnidarian Model” by Malamy and Shribak.
Michael Shribak, Marine Biological Laboratory/University of Chicago.
View Media
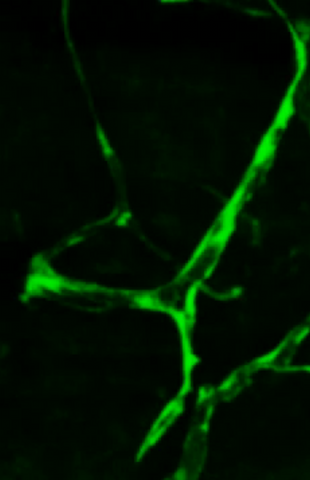
3405: Disrupted and restored vasculature development in frog embryos
3405: Disrupted and restored vasculature development in frog embryos
Disassembly of vasculature and reassembly after addition and then washout of 250 µM TBZ in kdr:GFP frogs. Related to images 3403 and 3404.
Hye Ji Cha, University of Texas at Austin
View Media
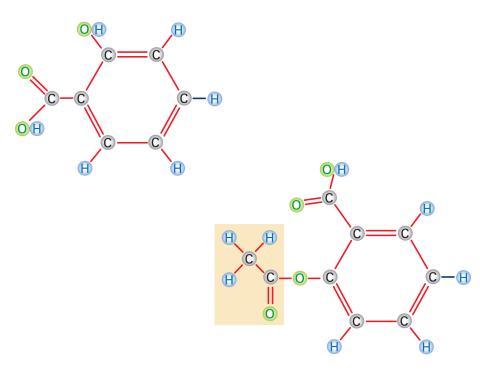
2529: Aspirin
2529: Aspirin
Acetylsalicylate (bottom) is the aspirin of today. Adding a chemical tag called an acetyl group (shaded box, bottom) to a molecule derived from willow bark (salicylate, top) makes the molecule less acidic (and easier on the lining of the digestive tract), but still effective at relieving pain. See image 2530 for a labeled version of this illustration. Featured in Medicines By Design.
Crabtree + Company
View Media
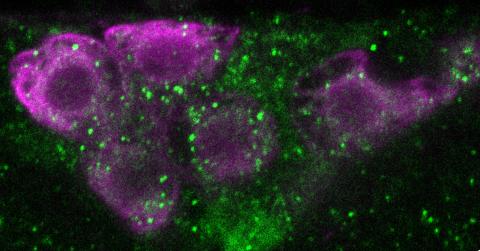
6982: Insulin production and fat sensing in fruit flies
6982: Insulin production and fat sensing in fruit flies
Fourteen neurons (magenta) in the adult Drosophila brain produce insulin, and fat tissue sends packets of lipids to the brain via the lipoprotein carriers (green). This image was captured using a confocal microscope and shows a maximum intensity projection of many slices.
Related to images 6983, 6984, and 6985.
Related to images 6983, 6984, and 6985.
Akhila Rajan, Fred Hutchinson Cancer Center
View Media
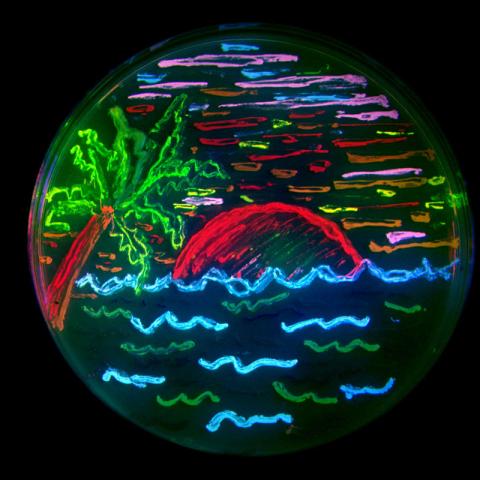
3492: Glowing bacteria make a pretty postcard
3492: Glowing bacteria make a pretty postcard
This tropical scene, reminiscent of a postcard from Key West, is actually a petri dish containing an artistic arrangement of genetically engineered bacteria. The image showcases eight of the fluorescent proteins created in the laboratory of the late Roger Y. Tsien, a cell biologist at the University of California, San Diego. Tsien, along with Osamu Shimomura of the Marine Biology Laboratory and Martin Chalfie of Columbia University, share the 2008 Nobel Prize in chemistry for their work on green fluorescent protein-a naturally glowing molecule from jellyfish that has become a powerful tool for studying molecules inside living cells.
Nathan C. Shaner, The Scintillon Institute
View Media
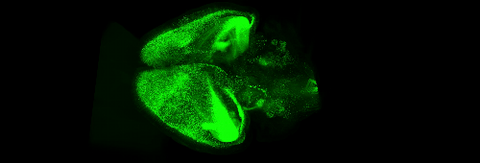
6931: Mouse brain 3
6931: Mouse brain 3
Various views of a mouse brain that was genetically modified so that subpopulations of its neurons glow. Researchers often study mice because they share many genes with people and can shed light on biological processes, development, and diseases in humans.
This video was captured using a light sheet microscope.
Related to images 6929 and 6930.
This video was captured using a light sheet microscope.
Related to images 6929 and 6930.
Prayag Murawala, MDI Biological Laboratory and Hannover Medical School.
View Media
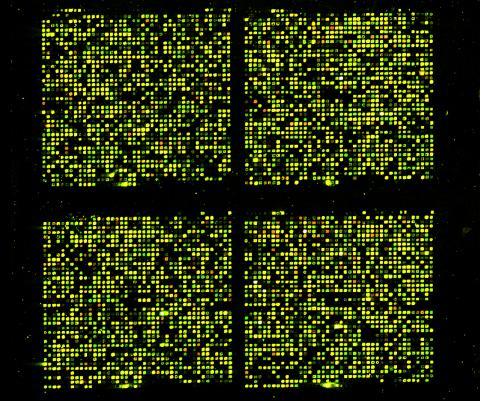
1070: Microarray 01
1070: Microarray 01
Microarrays, also called gene chips, are tools that let scientists track the activity of hundreds or thousands of genes simultaneously. For example, researchers can compare the activities of genes in healthy and diseased cells, allowing the scientists to pinpoint which genes and cell processes might be involved in the development of a disease.
Maggie Werner-Washburne, University of New Mexico, Albuquerque
View Media
1086: Natcher Building 06
1086: Natcher Building 06
NIGMS staff are located in the Natcher Building on the NIH campus.
Alisa Machalek, National Institute of General Medical Sciences
View Media
2358: Advanced Photon Source (APS) at Argonne National Lab
2358: Advanced Photon Source (APS) at Argonne National Lab
The intense X-rays produced by synchrotrons such as the Advanced Photon Source are ideally suited for protein structure determination. Using synchrotron X-rays and advanced computers scientists can determine protein structures at a pace unheard of decades ago.
Southeast Collaboratory for Structural Genomics
View Media

3424: White Poppy
3424: White Poppy
A white poppy. View cropped image of a poppy here 3423.
Judy Coyle, Donald Danforth Plant Science Center
View Media
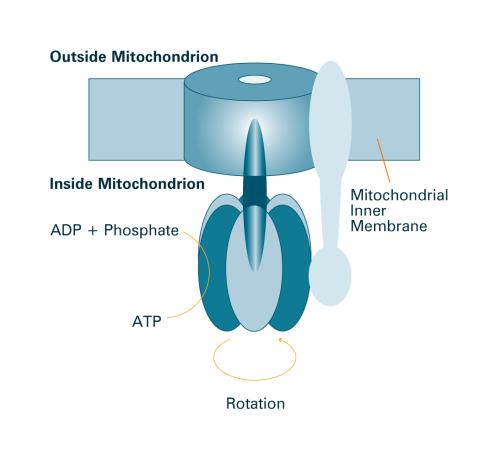
2518: ATP synthase (with labels)
2518: ATP synthase (with labels)
The world's smallest motor, ATP synthase, generates energy for the cell. See image 2517 for an unlabeled version of this illustration. Featured in The Chemistry of Health.
Crabtree + Company
View Media
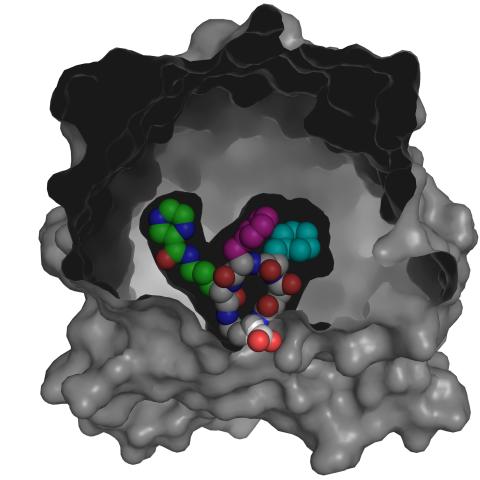
3416: X-ray co-crystal structure of Src kinase bound to a DNA-templated macrocycle inhibitor 4
3416: X-ray co-crystal structure of Src kinase bound to a DNA-templated macrocycle inhibitor 4
X-ray co-crystal structure of Src kinase bound to a DNA-templated macrocycle inhibitor. Related to 3413, 3414, 3415, 3417, 3418, and 3419.
Markus A. Seeliger, Stony Brook University Medical School and David R. Liu, Harvard University
View Media
3788: Yeast cells pack a punch
3788: Yeast cells pack a punch
Although they are tiny, microbes that are growing in confined spaces can generate a lot of pressure. In this video, yeast cells grow in a small chamber called a microfluidic bioreactor. As the cells multiply, they begin to bump into and squeeze each other, resulting in periodic bursts of cells moving into different parts of the chamber. The continually growing cells also generate a lot of pressure--the researchers conducting these experiments found that the pressure generated by the cells can be almost five times higher than that in a car tire--about 150 psi, or 10 times the atmospheric pressure. Occasionally, this pressure even caused the small reactor to burst. By tracking the growth of the yeast or other cells and measuring the mechanical forces generated, scientists can simulate microbial growth in various places such as water pumps, sewage lines or catheters to learn how damage to these devices can be prevented. To learn more how researchers used small bioreactors to gauge the pressure generated by growing microbes, see this press release from UC Berkeley.
Oskar Hallatschek, UC Berkeley
View Media
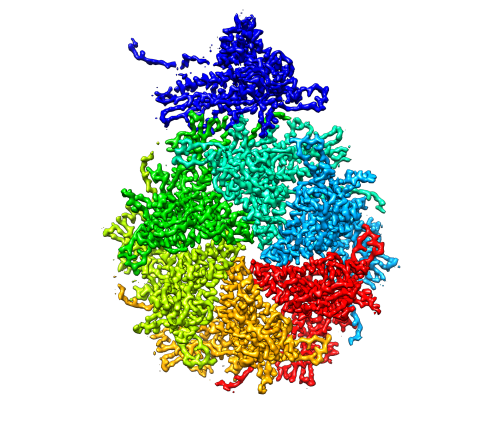
5875: Bacteriophage P22 capsid, detail
5875: Bacteriophage P22 capsid, detail
Detail of a subunit of the capsid, or outer cover, of bacteriophage P22, a virus that infects the Salmonella bacteria. Cryo-electron microscopy (cryo-EM) was used to capture details of the capsid proteins, each shown here in a separate color. Thousands of cryo-EM scans capture the structure and shape of all the individual proteins in the capsid and their position relative to other proteins. A computer model combines these scans into the image shown here. Related to image 5874.
Dr. Wah Chiu, Baylor College of Medicine
View Media
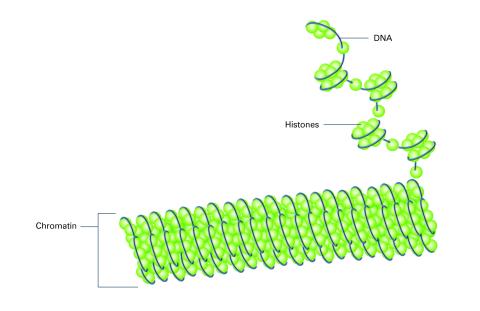
2561: Histones in chromatin (with labels)
2561: Histones in chromatin (with labels)
Histone proteins loop together with double-stranded DNA to form a structure that resembles beads on a string. See image 2560 for an unlabeled version of this illustration. Featured in The New Genetics.
Crabtree + Company
View Media
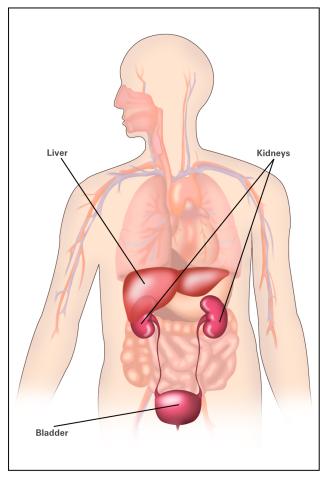
2497: Body toxins (with labels)
2497: Body toxins (with labels)
Body organs such as the liver and kidneys process chemicals and toxins. These "target" organs are susceptible to damage caused by these substances. See image 2496 for an unlabeled version of this illustration.
Crabtree + Company
View Media
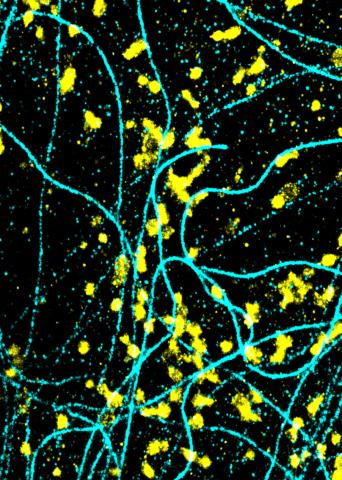
6889: Lysosomes and microtubules
6889: Lysosomes and microtubules
Lysosomes (yellow) and detyrosinated microtubules (light blue). Lysosomes are bubblelike organelles that take in molecules and use enzymes to break them down. Microtubules are strong, hollow fibers that provide structural support to cells. The researchers who took this image found that in epithelial cells, detyrosinated microtubules are a small subset of fibers, and they concentrate lysosomes around themselves. This image was captured using Stochastic Optical Reconstruction Microscopy (STORM).
Related to images 6890, 6891, and 6892.
Related to images 6890, 6891, and 6892.
Melike Lakadamyali, Perelman School of Medicine at the University of Pennsylvania.
View Media

2376: Protein purification facility
2376: Protein purification facility
The Center for Eukaryotic Structural Genomics protein purification facility is responsible for purifying all recombinant proteins produced by the center. The facility performs several purification steps, monitors the quality of the processes, and stores information about the biochemical properties of the purified proteins in the facility database.
Center for Eukaryotic Structural Genomics
View Media
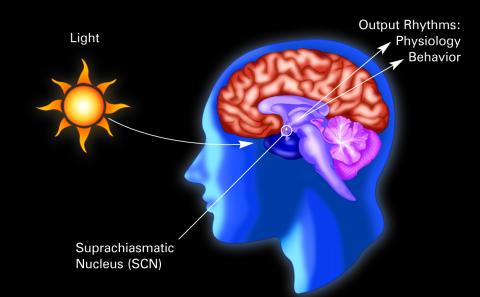
2569: Circadian rhythm (with labels)
2569: Circadian rhythm (with labels)
The human body keeps time with a master clock called the suprachiasmatic nucleus or SCN. Situated inside the brain, it's a tiny sliver of tissue about the size of a grain of rice, located behind the eyes. It sits quite close to the optic nerve, which controls vision, and this means that the SCN "clock" can keep track of day and night. The SCN helps control sleep and maintains our circadian rhythm--the regular, 24-hour (or so) cycle of ups and downs in our bodily processes such as hormone levels, blood pressure, and sleepiness. The SCN regulates our circadian rhythm by coordinating the actions of billions of miniature "clocks" throughout the body. These aren't actually clocks, but rather are ensembles of genes inside clusters of cells that switch on and off in a regular, 24-hour (or so) cycle in our physiological day.
Crabtree + Company
View Media
1087: Natcher Building 07
1087: Natcher Building 07
NIGMS staff are located in the Natcher Building on the NIH campus.
Alisa Machalek, National Institute of General Medical Sciences
View Media

2369: Protein purification robot in action 01
2369: Protein purification robot in action 01
A robot is transferring 96 purification columns to a vacuum manifold for subsequent purification procedures.
The Northeast Collaboratory for Structural Genomics
View Media
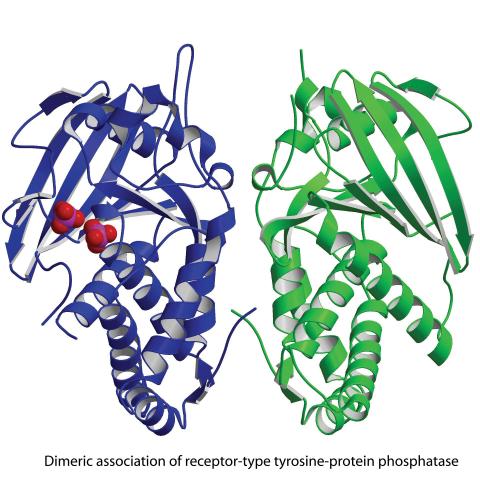
2349: Dimeric association of receptor-type tyrosine-protein phosphatase
2349: Dimeric association of receptor-type tyrosine-protein phosphatase
Model of the catalytic portion of an enzyme, receptor-type tyrosine-protein phosphatase from humans. The enzyme consists of two identical protein subunits, shown in blue and green. The groups made up of purple and red balls represent phosphate groups, chemical groups that can influence enzyme activity. This phosphatase removes phosphate groups from the enzyme tyrosine kinase, counteracting its effects.
New York Structural GenomiX Research Consortium, PSI
View Media

