Image and Video Gallery
This is a searchable collection of scientific photos, illustrations, and videos. The images and videos in this gallery are licensed under Creative Commons Attribution Non-Commercial ShareAlike 3.0. This license lets you remix, tweak, and build upon this work non-commercially, as long as you credit and license your new creations under identical terms.
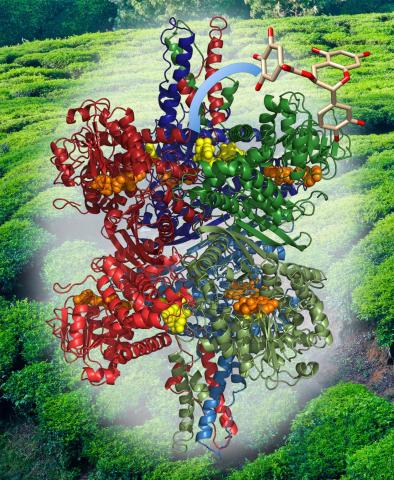
2571: VDAC video 02
Related to videos 2570 and 2572.
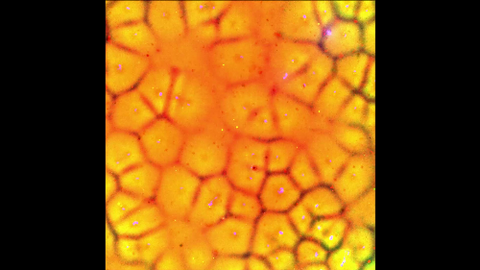
6587: Cell-like compartments emerging from scrambled frog eggs
For more photos of cell-like compartments from frog eggs view: 6584, 6585, 6586, 6591, 6592, and 6593.
For videos of cell-like compartments from frog eggs view: 6588, 6589, and 6590.
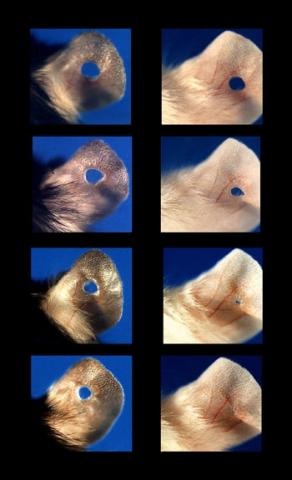
3426: Regeneration of Mouse Ears
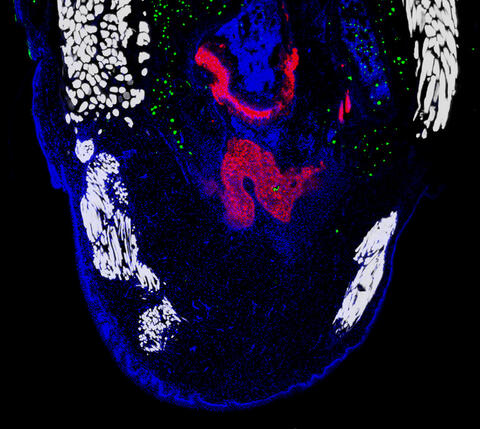
6968: Regenerating lizard tail

2361: Chromium X-ray source
6983: Genetic mosaicism in fruit flies
Related to images 6982, 6984, and 6985.
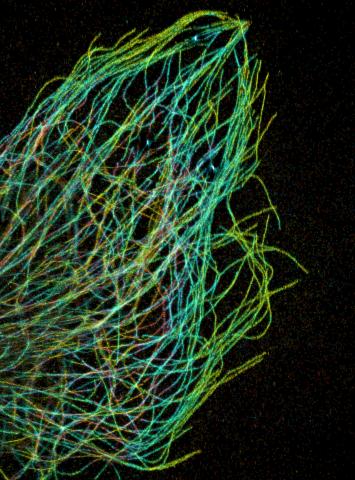
3611: Tiny strands of tubulin, a protein in a cell's skeleton
This image was part of the Life: Magnified exhibit that ran from June 3, 2014, to January 21, 2015, at Dulles International Airport.
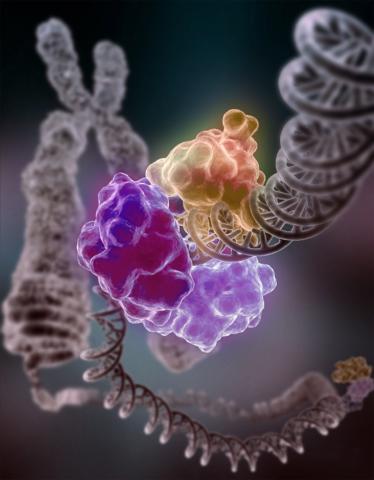
3493: Repairing DNA
6555: Floral pattern in a mixture of two bacterial species, Acinetobacter baylyi and Escherichia coli, grown on a semi-solid agar for 48 hours (photo 2)
See 6557 for a photo of this process at 24 hours on 0.75% agar surface.
See 6553 for another photo of this process at 48 hours on 1% agar surface.
See 6556 for a photo of this process at 72 hours on 0.5% agar surface.
See 6550 for a video of this process.
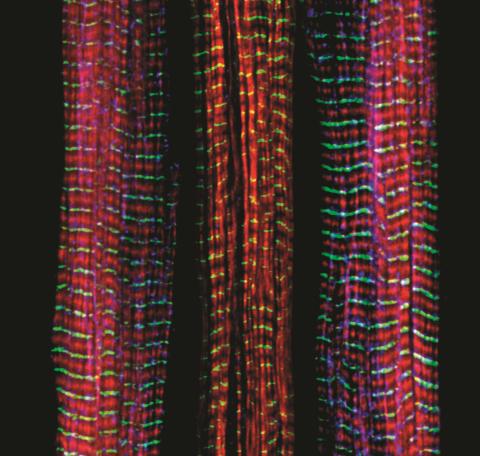
3630: Three muscle fibers; the middle has a defect found in some neuromuscular diseases
This image was part of the Life: Magnified exhibit that ran from June 3, 2014, to January 21, 2015, at Dulles International Airport.
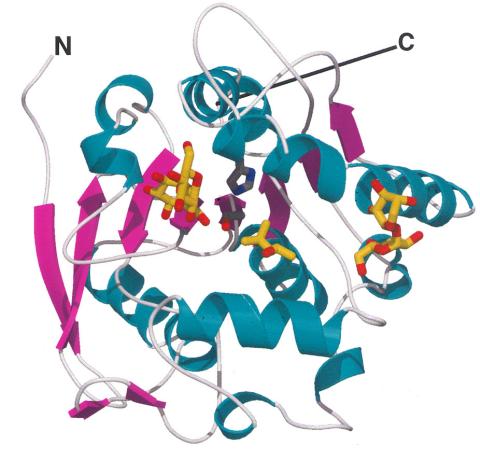
2378: Most abundant protein in M. tuberculosis
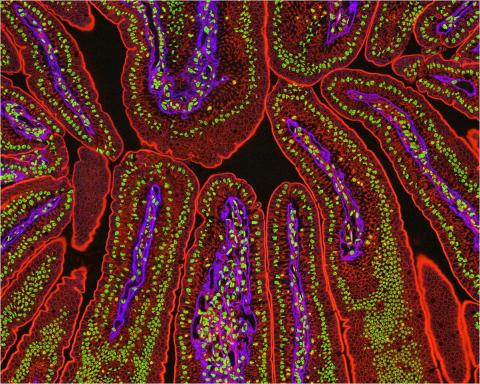
3390: NCMIR Intestine-2
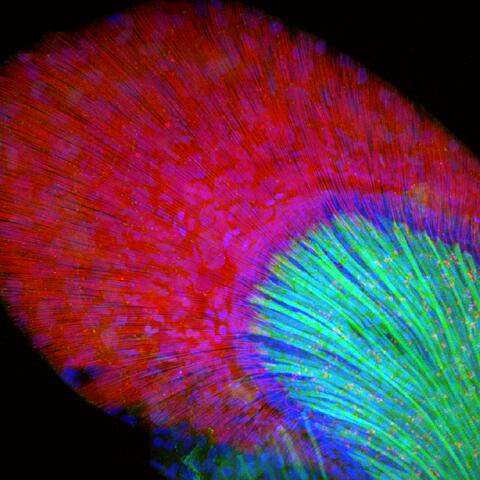
3598: Developing zebrafish fin
In this image, green fluorescent protein (GFP) is expressed where the gene sox9b is expressed. Collagen (red) marks the fin rays, and DNA, stained with a dye called DAPI, is in blue. sox9b plays many important roles during development, including the building of the heart and brain, and is also necessary for skeletal development. At the University of Wisconsin, researchers have found that exposure to contaminants that bind the aryl-hydrocarbon receptor results in the downregulation of sox9b. Loss of sox9b severely disrupts development in zebrafish and causes a life-threatening disorder called campomelic dysplasia (CD) in humans. CD is characterized by cardiovascular, neural, and skeletal defects. By studying the roles of genes such as sox9b in zebrafish, scientists hope to better understand normal development in humans as well as how to treat developmental disorders and diseases.
This image was part of the Life: Magnified exhibit that ran from June 3, 2014, to January 21, 2015, at Dulles International Airport.
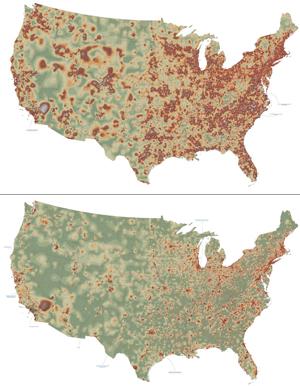
2320: Mapping disease spread

2409: Bacterial glucose isomerase
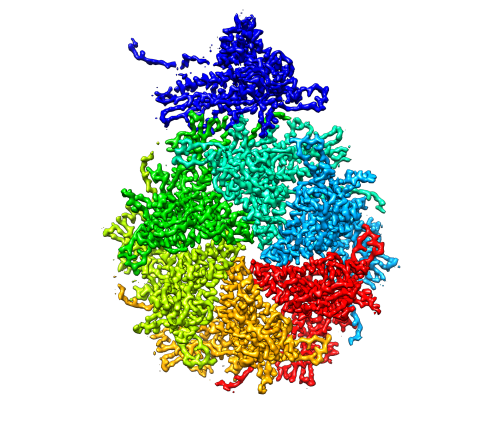
5875: Bacteriophage P22 capsid, detail
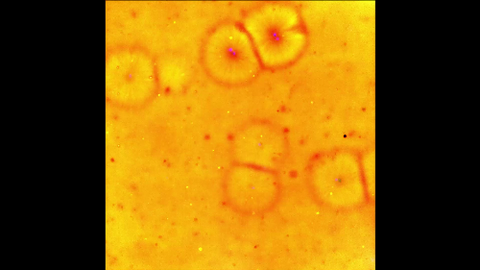
6588: Cell-like compartments emerging from scrambled frog eggs 2
For more photos of cell-like compartments from frog eggs view: 6584, 6585, 6586, 6591, 6592, and 6593.
For videos of cell-like compartments from frog eggs view: 6587, 6589, and 6590.
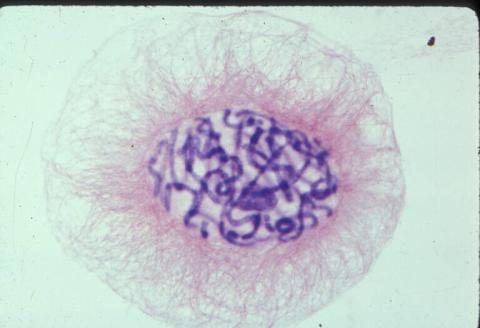
1014: Lily mitosis 04
Related to images 1010, 1011, 1012, 1013, 1015, 1016, 1017, 1018, 1019, and 1021.
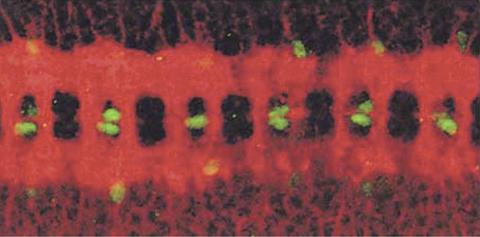
1091: Nerve and glial cells in fruit fly embryo

2345: Magnesium transporter protein from E. faecalis

5895: Bioluminescence in a Tube
From Biomedical Beat article July 2017: Chasing Fireflies—and Better Cellular Imaging Techniques

3272: Ear hair cells derived from embryonic stem cells
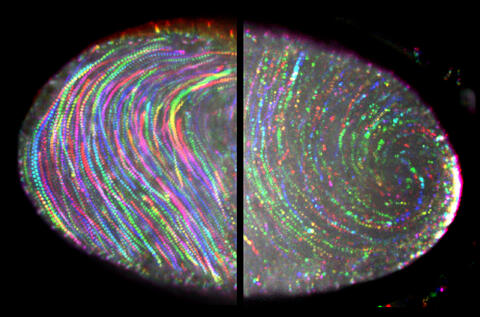
6809: Fruit fly egg ooplasmic streaming
More information on the research that produced this image can be found in the Journal of Cell Biology paper “Ooplasmic flow cooperates with transport and anchorage in Drosophila oocyte posterior determination” by Lu et al.
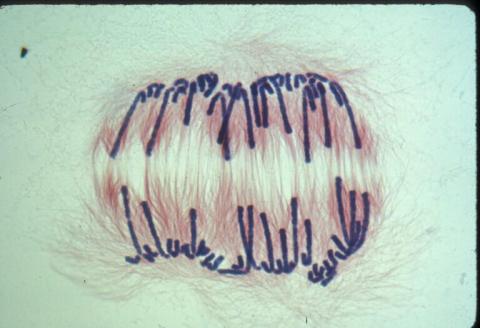
1010: Lily mitosis 10
Related to images 1011, 1012, 1013, 1014, 1015, 1016, 1017, 1018, 1019, and 1021.

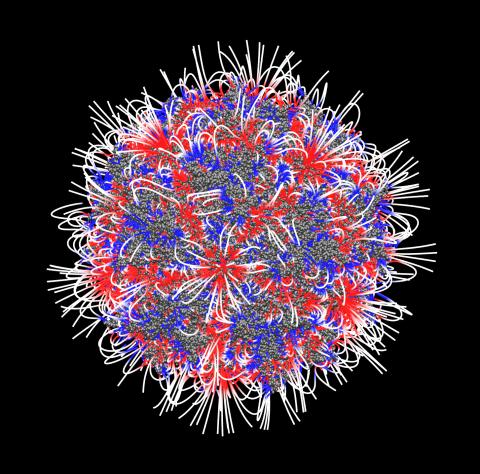
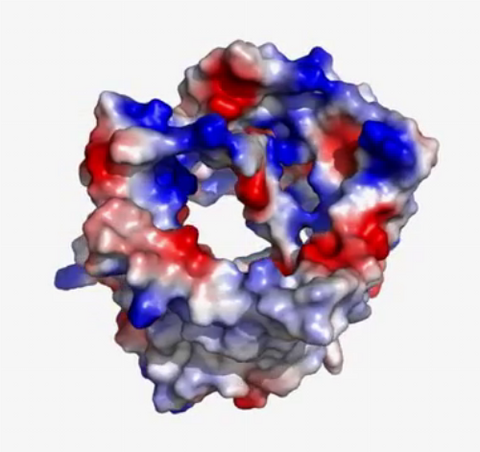
3374: Electrostatic map of the adeno-associated virus
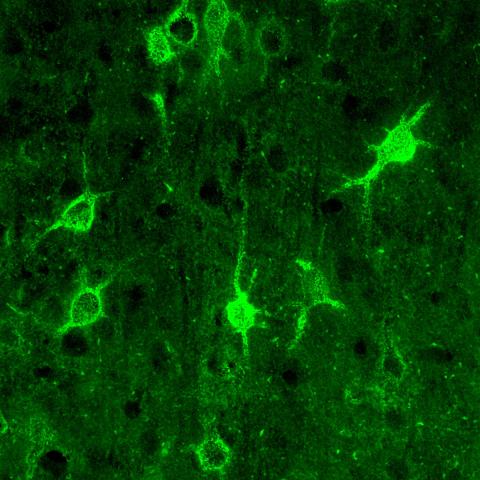
3741: Confocal microscopy of perineuronal nets in the brain 1
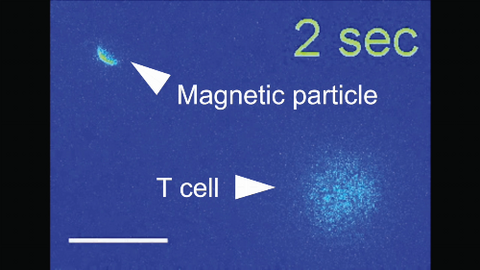
6800: Magnetic Janus particle activating a T cell
More details can be found in the Angewandte Chemie paper “Remote control of T cell activation using magnetic Janus particles” by Lee et al. This video was captured using epi-fluorescence microscopy.
Related to video 6801.
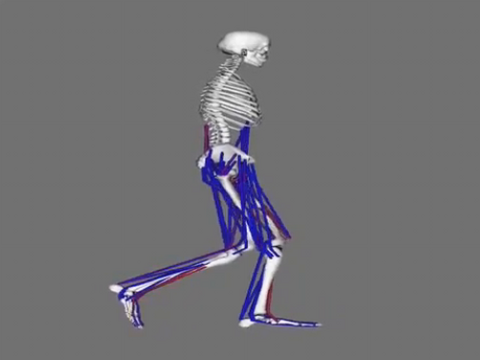
6598: Simulation of leg muscles moving
2307: Cells frozen in time
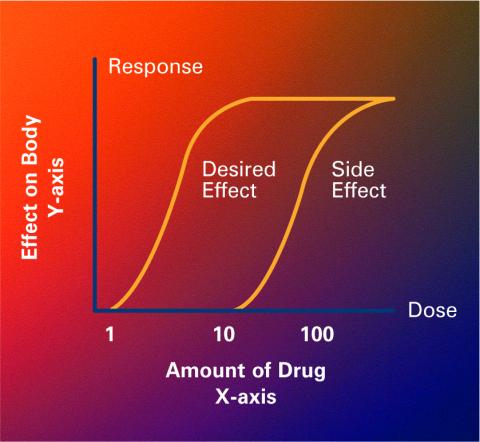
2533: Dose response curves
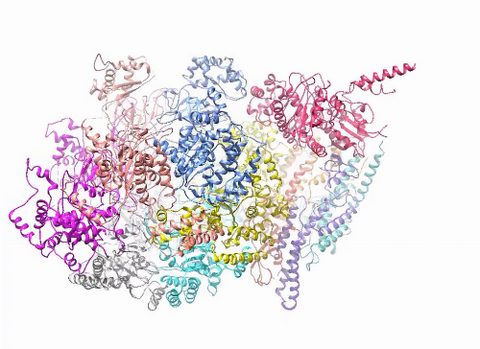
3750: A dynamic model of the DNA helicase protein complex
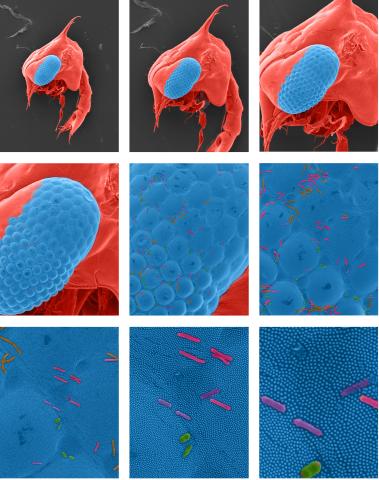
1251: Crab larva eye
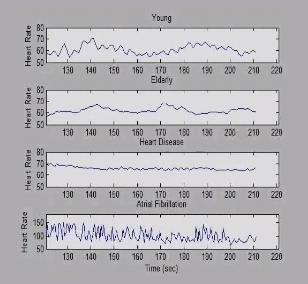
3596: Heart rates time series image
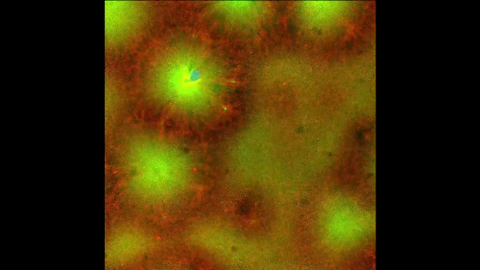
6590: Cell-like compartments emerging from scrambled frog eggs 4
For more photos of cell-like compartments from frog eggs view: 6584, 6585, 6586, 6591, 6592, and 6593.
For videos of cell-like compartments from frog eggs view: 6587, 6588, 6589.
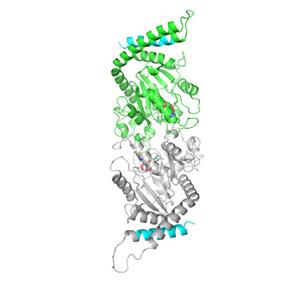
2744: Dynamin structure
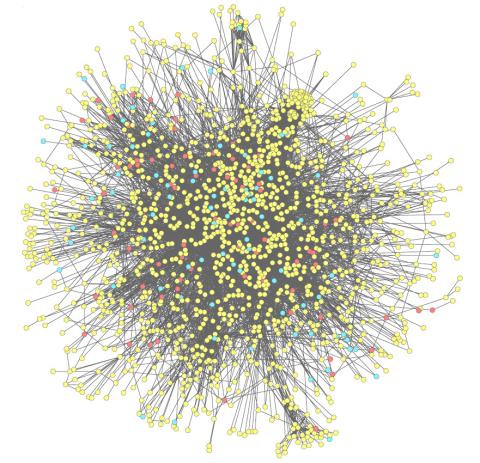
2743: Molecular interactions
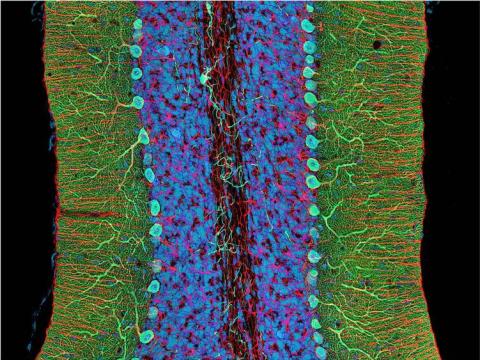
3371: Mouse cerebellum close-up
This image was part of the Life: Magnified exhibit that ran from June 3, 2014, to January 21, 2015, at Dulles International Airport.
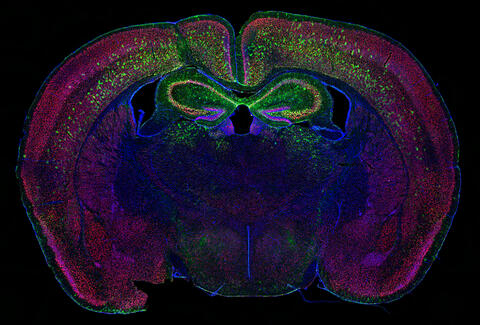
6780: Calling Cards in a mouse brain
Related to video
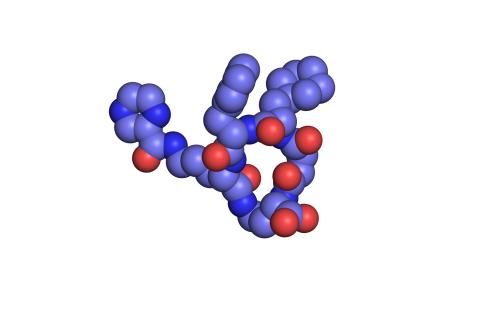
3413: X-ray co-crystal structure of Src kinase bound to a DNA-templated macrocycle inhibitor 1
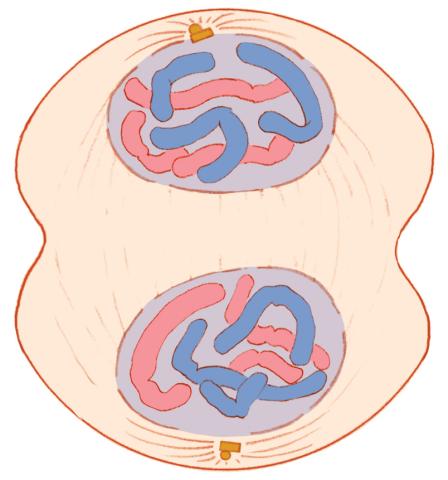
1332: Mitosis - telophase
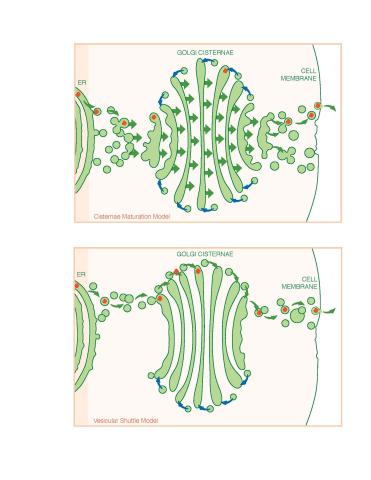
1278: Golgi theories
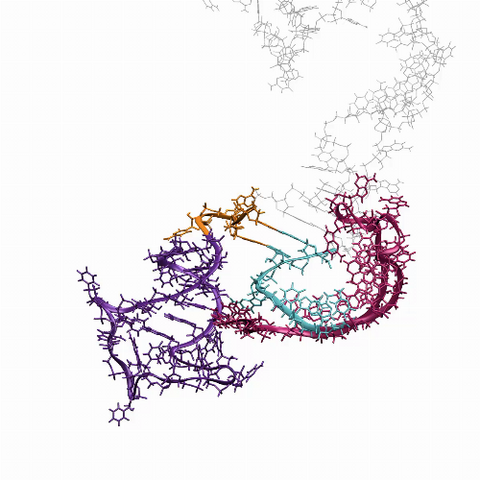
6625: RNA folding in action
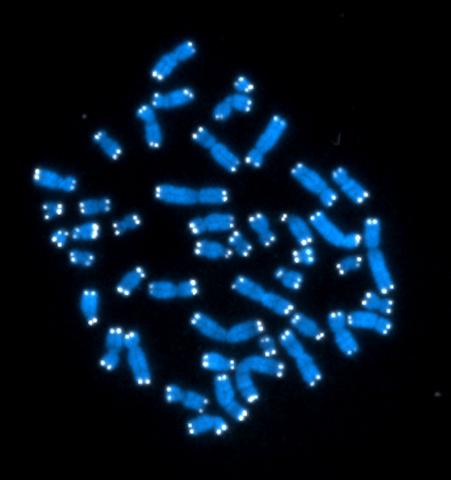
2626: Telomeres
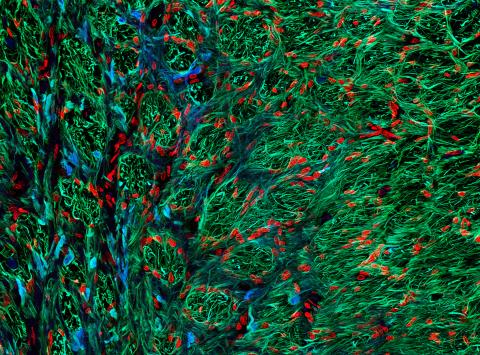
5852: Optic nerve astrocytes

2305: Beaded bacteriophage
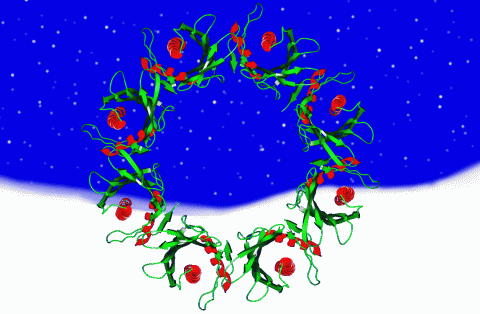
2372: Wreath-shaped protein from X. campestris
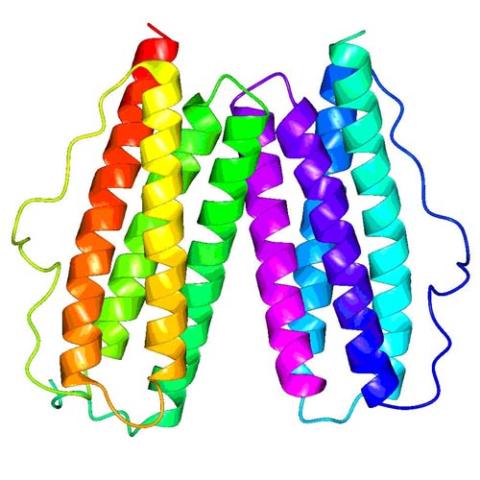
2343: Protein rv2844 from M. tuberculosis
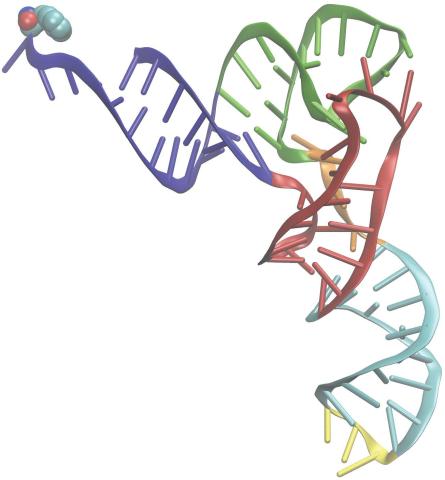
3406: Phenylalanine tRNA molecule