Switch to List View
Image and Video Gallery
This is a searchable collection of scientific photos, illustrations, and videos. The images and videos in this gallery are licensed under Creative Commons Attribution Non-Commercial ShareAlike 3.0. This license lets you remix, tweak, and build upon this work non-commercially, as long as you credit and license your new creations under identical terms.
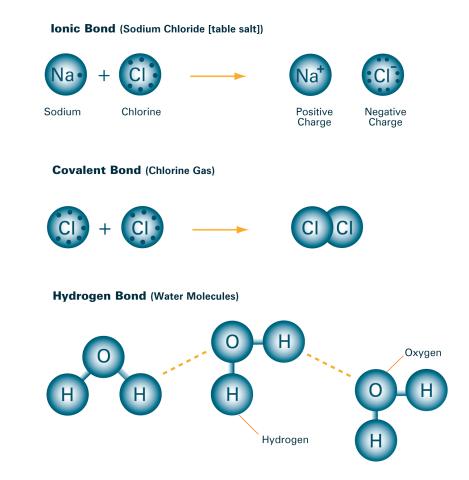
2520: Bond types (with labels)
2520: Bond types (with labels)
Ionic and covalent bonds hold molecules, like sodium chloride and chlorine gas, together. Hydrogen bonds among molecules, notably involving water, also play an important role in biology. See image 2519 for an unlabeled version of this illustration. Featured in The Chemistry of Health.
Crabtree + Company
View Media
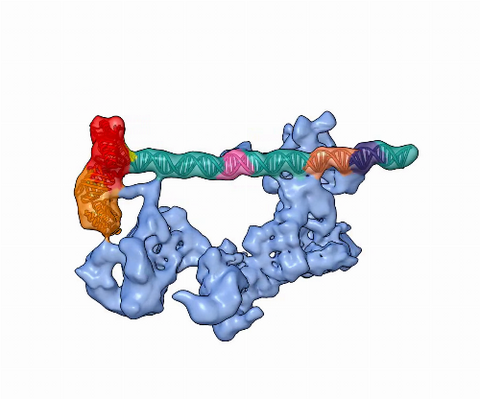
5730: Dynamic cryo-EM model of the human transcription preinitiation complex
5730: Dynamic cryo-EM model of the human transcription preinitiation complex
Gene transcription is a process by which information encoded in DNA is transcribed into RNA. It's essential for all life and requires the activity of proteins, called transcription factors, that detect where in a DNA strand transcription should start. In eukaryotes (i.e., those that have a nucleus and mitochondria), a protein complex comprising 14 different proteins is responsible for sniffing out transcription start sites and starting the process. This complex represents the core machinery to which an enzyme, named RNA polymerase, can bind to and read the DNA and transcribe it to RNA. Scientists have used cryo-electron microscopy (cryo-EM) to visualize the TFIID-RNA polymerase-DNA complex in unprecedented detail. This animation shows the different TFIID components as they contact DNA and recruit the RNA polymerase for gene transcription.
To learn more about the research that has shed new light on gene transcription, see this news release from Berkeley Lab.
Related to image 3766.
To learn more about the research that has shed new light on gene transcription, see this news release from Berkeley Lab.
Related to image 3766.
Eva Nogales, Berkeley Lab
View Media
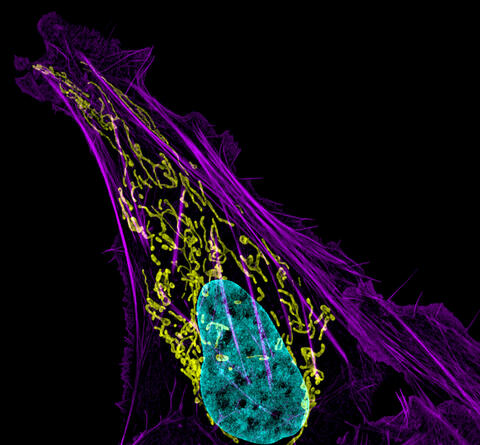
3626: Bone cancer cell
3626: Bone cancer cell
This image shows an osteosarcoma cell with DNA in blue, energy factories (mitochondria) in yellow, and actin filaments—part of the cellular skeleton—in purple. One of the few cancers that originate in the bones, osteosarcoma is rare, with about a thousand new cases diagnosed each year in the United States.
This image was part of the Life: Magnified exhibit that ran from June 3, 2014, to January 21, 2015, at Dulles International Airport.
This image was part of the Life: Magnified exhibit that ran from June 3, 2014, to January 21, 2015, at Dulles International Airport.
Dylan Burnette and Jennifer Lippincott-Schwartz, NICHD
View Media
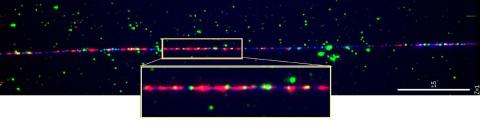
2475: Chromosome fiber 01
2475: Chromosome fiber 01
This microscopic image shows a chromatin fiber--a DNA molecule bound to naturally occurring proteins.
Marc Green and Susan Forsburg, University of Southern California
View Media

1335: Telomerase illustration
1335: Telomerase illustration
Reactivating telomerase in our cells does not appear to be a good way to extend the human lifespan. Cancer cells reactivate telomerase.
Judith Stoffer
View Media
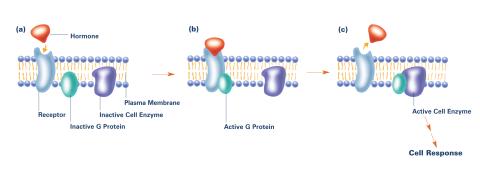
2538: G switch (with labels and stages)
2538: G switch (with labels and stages)
The G switch allows our bodies to respond rapidly to hormones. G proteins act like relay batons to pass messages from circulating hormones into cells. A hormone (red) encounters a receptor (blue) in the membrane of a cell. Next, a G protein (green) becomes activated and makes contact with the receptor to which the hormone is attached. Finally, the G protein passes the hormone's message to the cell by switching on a cell enzyme (purple) that triggers a response. See image 2536 and 2537 for other versions of this image. Featured in Medicines By Design.
Crabtree + Company
View Media

7011: Hawaiian bobtail squid
7011: Hawaiian bobtail squid
An adult Hawaiian bobtail squid, Euprymna scolopes, swimming next to a submerged hand.
Related to image 7010 and video 7012.
Related to image 7010 and video 7012.
Margaret J. McFall-Ngai, Carnegie Institution for Science/California Institute of Technology, and Edward G. Ruby, California Institute of Technology.
View Media
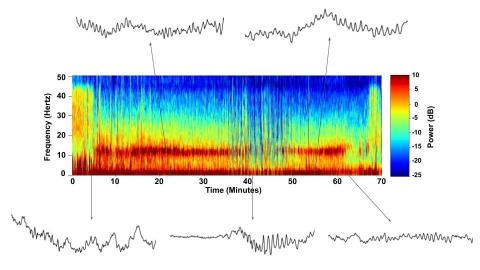
6779: Brain waves of a patient anesthetized with propofol
6779: Brain waves of a patient anesthetized with propofol
A representation of a patient’s brain waves after receiving the anesthetic propofol. All anesthetics create brain wave changes that vary depending on the patient’s age and the type and dose of anesthetic used. These changes are visible in raw electroencephalogram (EEG) readings, but they’re easier to interpret using a spectrogram where the signals are broken down by time (x-axis), frequency (y-axis), and power (color scale). This spectrogram shows the changes in brain waves before, during, and after propofol-induced anesthesia. The patient is unconscious from minute 5, upon propofol administration, through minute 69 (change in power and frequency). But, between minutes 35 and 48, the patient fell into a profound state of unconsciousness (disappearance of dark red oscillations between 8 to 12 Hz), which required the anesthesiologist to adjust the rate of propofol administration. The propofol was stopped at minute 62 and the patient woke up around minute 69.
Emery N. Brown, M.D., Ph.D., Massachusetts General Hospital/Harvard Medical School, Picower Institute for Learning and Memory, and Massachusetts Institute of Technology.
View Media
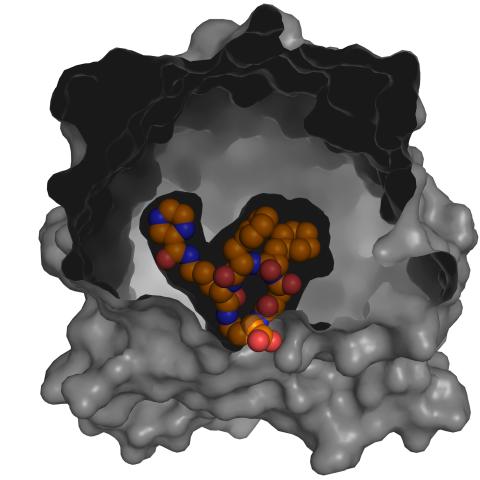
3417: X-ray co-crystal structure of Src kinase bound to a DNA-templated macrocycle inhibitor 5
3417: X-ray co-crystal structure of Src kinase bound to a DNA-templated macrocycle inhibitor 5
X-ray co-crystal structure of Src kinase bound to a DNA-templated macrocycle inhibitor. Related to images 3413, 3414, 3415, 3416, 3418, and 3419.
Markus A. Seeliger, Stony Brook University Medical School and David R. Liu, Harvard University
View Media
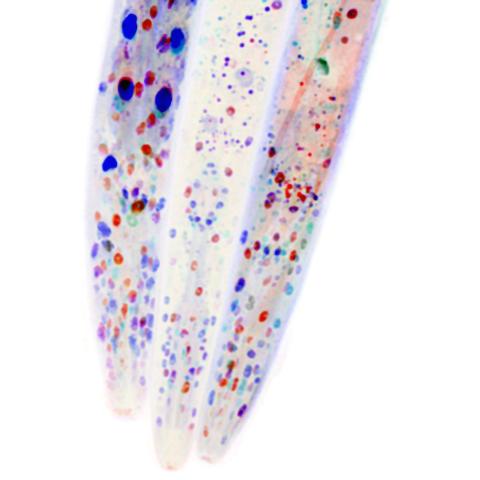
6534: Mosaicism in C. elegans (White Background)
6534: Mosaicism in C. elegans (White Background)
In the worm C. elegans, double-stranded RNA made in neurons can silence matching genes in a variety of cell types through the transport of RNA between cells. The head region of three worms that were genetically modified to express a fluorescent protein were imaged and the images were color-coded based on depth. The worm on the left lacks neuronal double-stranded RNA and thus every cell is fluorescent. In the middle worm, the expression of the fluorescent protein is silenced by neuronal double-stranded RNA and thus most cells are not fluorescent. The worm on the right lacks an enzyme that amplifies RNA for silencing. Surprisingly, the identities of the cells that depend on this enzyme for gene silencing are unpredictable. As a result, worms of identical genotype are nevertheless random mosaics for how the function of gene silencing is carried out. For more, see journal article and press release. Related to image 6532.
Snusha Ravikumar, Ph.D., University of Maryland, College Park, and Antony M. Jose, Ph.D., University of Maryland, College Park
View Media
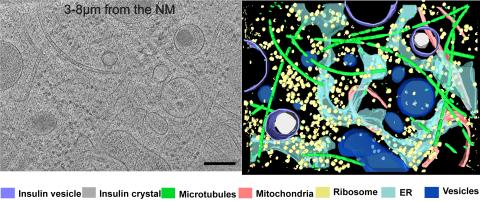
6608: Cryo-ET cross-section of a rat pancreas cell
6608: Cryo-ET cross-section of a rat pancreas cell
On the left, a cross-section slice of a rat pancreas cell captured using cryo-electron tomography (cryo-ET). On the right, a 3D, color-coded version of the image highlighting cell structures. Visible features include microtubules (neon-green rods), ribosomes (small yellow circles), and vesicles (dark-blue circles). These features are surrounded by the partially visible endoplasmic reticulum (light blue). The black line at the bottom right of the left image represents 200 nm. Related to image 6607.
Xianjun Zhang, University of Southern California.
View Media
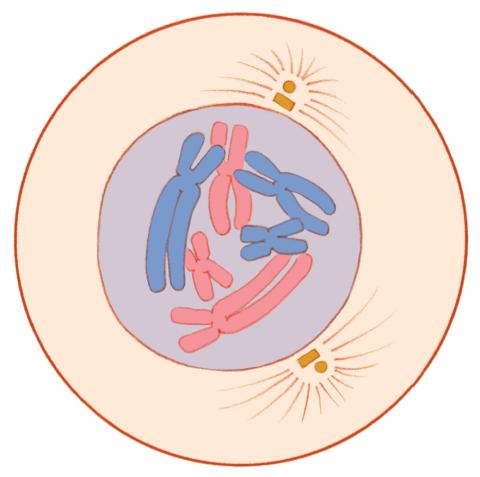
1330: Mitosis - prophase
1330: Mitosis - prophase
A cell in prophase, near the start of mitosis: In the nucleus, chromosomes condense and become visible. In the cytoplasm, the spindle forms. Mitosis is responsible for growth and development, as well as for replacing injured or worn out cells throughout the body. For simplicity, mitosis is illustrated here with only six chromosomes.
Judith Stoffer
View Media
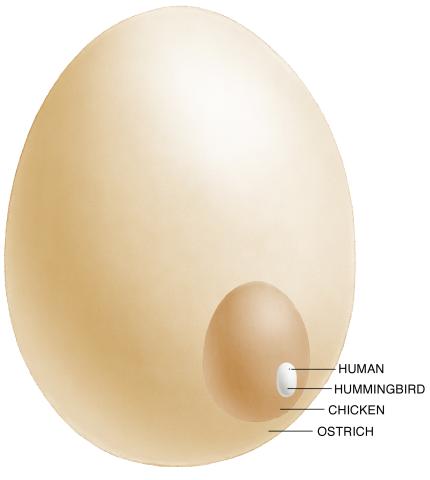
1339: Egg comparison
1339: Egg comparison
The largest human cell (by volume) is the egg. Human eggs are 150 micrometers in diameter and you can just barely see one with a naked eye. In comparison, consider the eggs of chickens...or ostriches!
Judith Stoffer
View Media
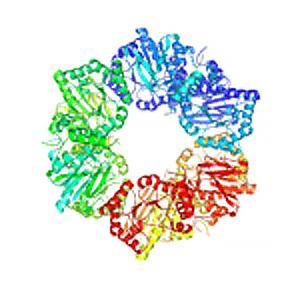
2355: Nicotinic acid phosphoribosyltransferase
2355: Nicotinic acid phosphoribosyltransferase
Model of the enzyme nicotinic acid phosphoribosyltransferase. This enzyme, from the archaebacterium, Pyrococcus furiosus, is expected to be structurally similar to a clinically important human protein called B-cell colony enhancing factor based on amino acid sequence similarities and structure prediction methods. The structure consists of identical protein subunits, each shown in a different color, arranged in a ring.
Berkeley Structural Genomics Center, PSI
View Media
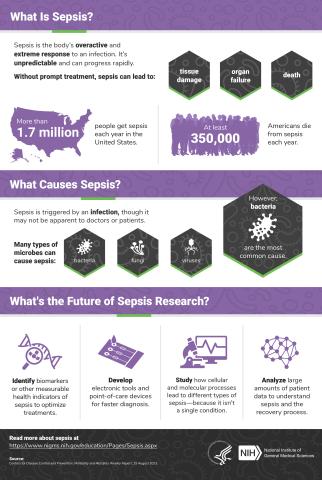
6536: Sepsis Infographic
6536: Sepsis Infographic
Sepsis is the body’s overactive and extreme response to an infection. More than 1.7 million people get sepsis each year in the United States. Without prompt treatment, sepsis can lead to tissue damage, organ failure, and death. Many NIGMS-supported researchers are working to improve sepsis diagnosis and treatment. Learn more with our sepsis featured topic page.
See 6551 for the Spanish version of this infographic.
See 6551 for the Spanish version of this infographic.
National Institute of General Medical Sciences
View Media
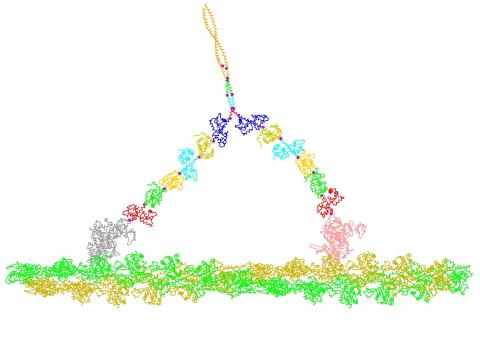
2754: Myosin V binding to actin
2754: Myosin V binding to actin
This simulation of myosin V binding to actin was created using the software tool Protein Mechanica. With Protein Mechanica, researchers can construct models using information from a variety of sources: crystallography, cryo-EM, secondary structure descriptions, as well as user-defined solid shapes, such as spheres and cylinders. The goal is to enable experimentalists to quickly and easily simulate how different parts of a molecule interact.
Simbios, NIH Center for Biomedical Computation at Stanford
View Media
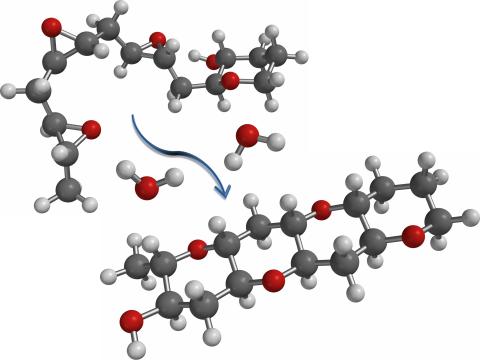
2490: Cascade reaction promoted by water
2490: Cascade reaction promoted by water
This illustration of an epoxide-opening cascade promoted by water emulates the proposed biosynthesis of some of the Red Tide toxins.
Tim Jamison, Massachusetts Institute of Technology
View Media

2531: Drugs enter skin
2531: Drugs enter skin
Drugs enter different layers of skin via intramuscular, subcutaneous, or transdermal delivery methods. See image 2532 for a labeled version of this illustration. Featured in Medicines By Design.
Crabtree + Company
View Media

6902: Arachnoidiscus diatom
6902: Arachnoidiscus diatom
An Arachnoidiscus diatom with a diameter of 190µm. Diatoms are microscopic algae that have cell walls made of silica, which is the strongest known biological material relative to its density. In Arachnoidiscus, the cell wall is a radially symmetric pillbox-like shell composed of overlapping halves that contain intricate and delicate patterns. Sometimes, Arachnoidiscus is called “a wheel of glass.”
This image was taken with the orientation-independent differential interference contrast microscope.
This image was taken with the orientation-independent differential interference contrast microscope.
Michael Shribak, Marine Biological Laboratory/University of Chicago.
View Media

6887: Chromatin in human fibroblast
6887: Chromatin in human fibroblast
The nucleus of a human fibroblast cell with chromatin—a substance made up of DNA and proteins—shown in various colors. Fibroblasts are one of the most common types of cells in mammalian connective tissue, and they play a key role in wound healing and tissue repair. This image was captured using Stochastic Optical Reconstruction Microscopy (STORM).
Related to images 6888 and 6893.
Related to images 6888 and 6893.
Melike Lakadamyali, Perelman School of Medicine at the University of Pennsylvania.
View Media
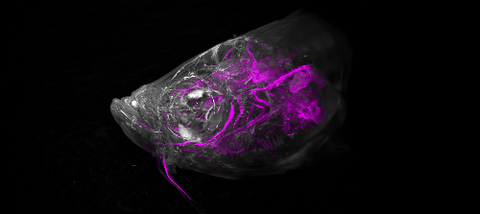
6933: Zebrafish head vasculature video
6933: Zebrafish head vasculature video
Various views of a zebrafish head with blood vessels shown in purple. Researchers often study zebrafish because they share many genes with humans, grow and reproduce quickly, and have see-through eggs and embryos, which make it easy to study early stages of development.
This video was captured using a light sheet microscope.
Related to image 6934.
This video was captured using a light sheet microscope.
Related to image 6934.
Prayag Murawala, MDI Biological Laboratory and Hannover Medical School.
View Media
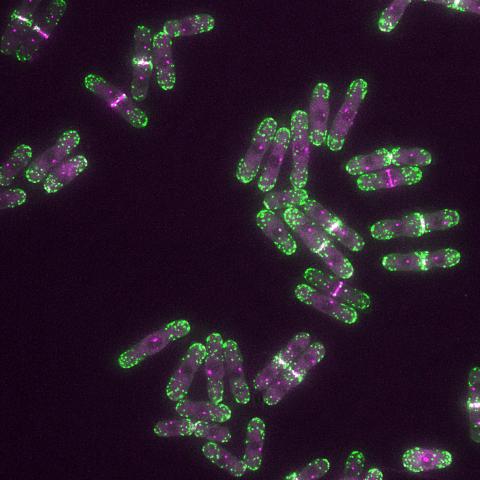
6793: Yeast cells with endocytic actin patches
6793: Yeast cells with endocytic actin patches
Yeast cells with endocytic actin patches (green). These patches help cells take in outside material. When a cell is in interphase, patches concentrate at its ends. During later stages of cell division, patches move to where the cell splits. This image was captured using wide-field microscopy with deconvolution.
Related to images 6791, 6792, 6794, 6797, 6798, and videos 6795 and 6796.
Related to images 6791, 6792, 6794, 6797, 6798, and videos 6795 and 6796.
Alaina Willet, Kathy Gould’s lab, Vanderbilt University.
View Media
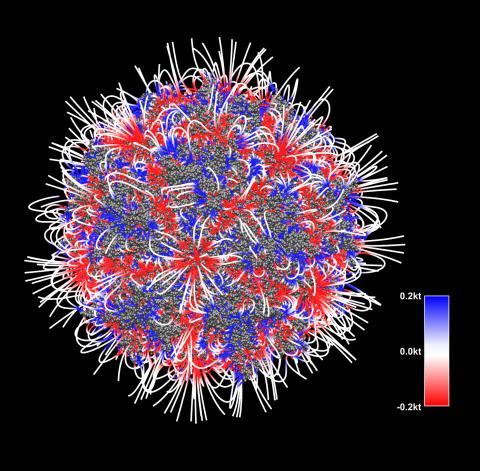
3375: Electrostatic map of the adeno-associated virus with scale
3375: Electrostatic map of the adeno-associated virus with scale
The new highly efficient parallelized DelPhi software was used to calculate the potential map distribution of an entire virus, the adeno-associated virus, which is made up of more than 484,000 atoms. Despite the relatively large dimension of this biological system, resulting in 815x815x815 mesh points, the parallelized DelPhi, utilizing 100 CPUs, completed the calculations within less than three minutes. Related to image 3374.
Emil Alexov, Clemson University
View Media

2313: Colorful communication
2313: Colorful communication
The marine bacterium Vibrio harveyi glows when near its kind. This luminescence, which results from biochemical reactions, is part of the chemical communication used by the organisms to assess their own population size and distinguish themselves from other types of bacteria. But V. harveyi only light up when part of a large group. This communication, called quorum sensing, speaks for itself here on a lab dish, where more densely packed areas of the bacteria show up blue. Other types of bacteria use quorum sensing to release toxins, trigger disease, and evade the immune system.
Bonnie Bassler, Princeton University
View Media

3327: Diversity oriented synthesis: generating skeletal diversity using folding processes
3327: Diversity oriented synthesis: generating skeletal diversity using folding processes
This 1 1/2-minute video animation was produced for chemical biologist Stuart Schreiber's lab page. The animation shows how diverse chemical structures can be produced in the lab.
Eric Keller
View Media
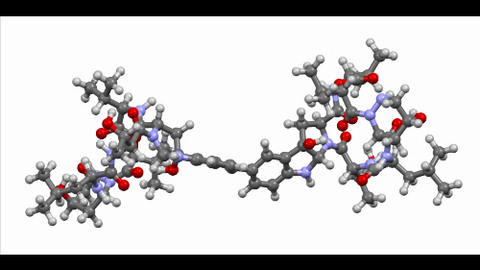
6851: Himastatin, 360-degree view
6851: Himastatin, 360-degree view
A 360-degree view of the molecule himastatin, which was first isolated from the bacterium Streptomyces himastatinicus. Himastatin shows antibiotic activity. The researchers who created this video developed a new, more concise way to synthesize himastatin so it can be studied more easily.
More information about the research that produced this video can be found in the Science paper “Total synthesis of himastatin” by D’Angelo et al.
Related to images 6848 and 6850.
More information about the research that produced this video can be found in the Science paper “Total synthesis of himastatin” by D’Angelo et al.
Related to images 6848 and 6850.
Mohammad Movassaghi, Massachusetts Institute of Technology.
View Media
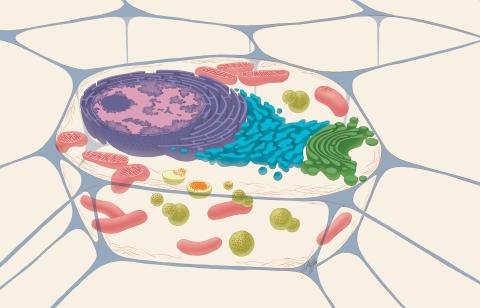
1274: Animal cell
1274: Animal cell
A typical animal cell, sliced open to reveal a cross-section of organelles.
Judith Stoffer
View Media
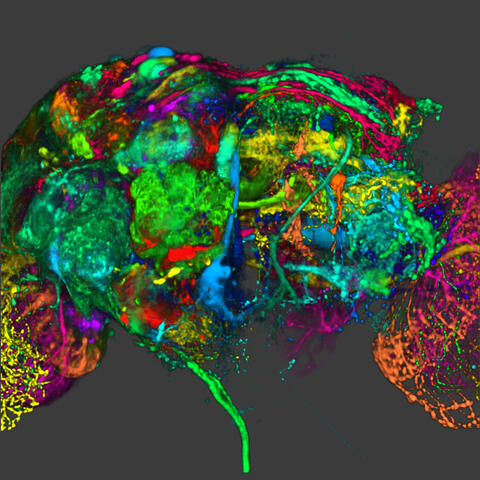
5838: Color coding of the Drosophila brain - image
5838: Color coding of the Drosophila brain - image
This image results from a research project to visualize which regions of the adult fruit fly (Drosophila) brain derive from each neural stem cell. First, researchers collected several thousand fruit fly larvae and fluorescently stained a random stem cell in the brain of each. The idea was to create a population of larvae in which each of the 100 or so neural stem cells was labeled at least once. When the larvae grew to adults, the researchers examined the flies’ brains using confocal microscopy. With this technique, the part of a fly’s brain that derived from a single, labeled stem cell “lights up. The scientists photographed each brain and digitally colorized its lit-up area. By combining thousands of such photos, they created a three-dimensional, color-coded map that shows which part of the Drosophila brain comes from each of its ~100 neural stem cells. In other words, each colored region shows which neurons are the progeny or “clones” of a single stem cell. This work established a hierarchical structure as well as nomenclature for the neurons in the Drosophila brain. Further research will relate functions to structures of the brain.
Related to image 5868 and video 5843
Related to image 5868 and video 5843
Yong Wan from Charles Hansen’s lab, University of Utah. Data preparation and visualization by Masayoshi Ito in the lab of Kei Ito, University of Tokyo.
View Media
1058: Lily mitosis 01
1058: Lily mitosis 01
A light microscope image shows the chromosomes, stained dark blue, in a dividing cell of an African globe lily (Scadoxus katherinae). This is one frame of a time-lapse sequence that shows cell division in action. The lily is considered a good organism for studying cell division because its chromosomes are much thicker and easier to see than human ones.
Andrew S. Bajer, University of Oregon, Eugene
View Media
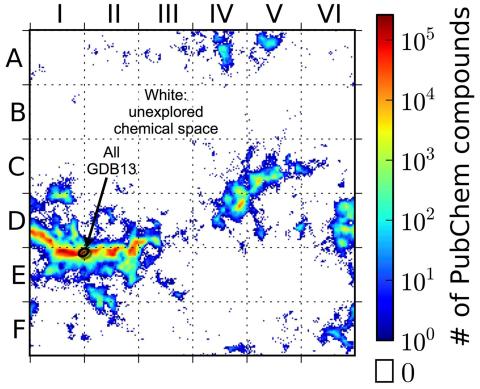
3458: Computer algorithm
3458: Computer algorithm
This computer algorithm plots all feasible small carbon-based molecules as though they were cities on a map and identifies huge, unexplored spaces that may help fuel research into new drug therapies. Featured in the May 16, 2013 issue of Biomedical Beat.
Aaron Virshup, Julia Contreras-Garcia, Peter Wipf, Weitao Yang and David Beratan, University of Pittsburgh Center for Chemical Methodologies and Library Development
View Media

1334: Aging book of life
1334: Aging book of life
Damage to each person's genome, often called the "Book of Life," accumulates with time. Such DNA mutations arise from errors in the DNA copying process, as well as from external sources, such as sunlight and cigarette smoke. DNA mutations are known to cause cancer and also may contribute to cellular aging.
Judith Stoffer
View Media
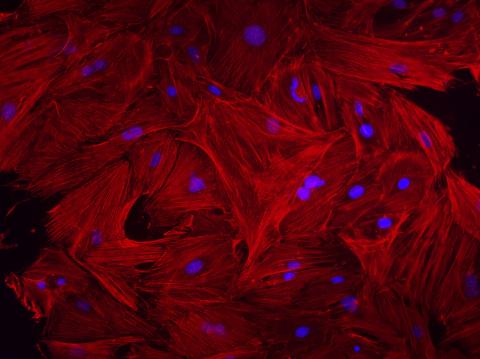
3281: Mouse heart fibroblasts
3281: Mouse heart fibroblasts
This image shows mouse fetal heart fibroblast cells. The muscle protein actin is stained red, and the cell nuclei are stained blue. The image was part of a study investigating stem cell-based approaches to repairing tissue damage after a heart attack. Image and caption information courtesy of the California Institute for Regenerative Medicine.
Kara McCloskey lab, University of California, Merced, via CIRM
View Media

3658: Electrostatic map of human spermine synthase
3658: Electrostatic map of human spermine synthase
From PDB entry 3c6k, Crystal structure of human spermine synthase in complex with spermidine and 5-methylthioadenosine.
Emil Alexov, Clemson University
View Media

1287: Mitochondria
1287: Mitochondria
Bean-shaped mitochondria are cells' power plants. These organelles have their own DNA and replicate independently. The highly folded inner membranes are the site of energy generation.
Judith Stoffer
View Media
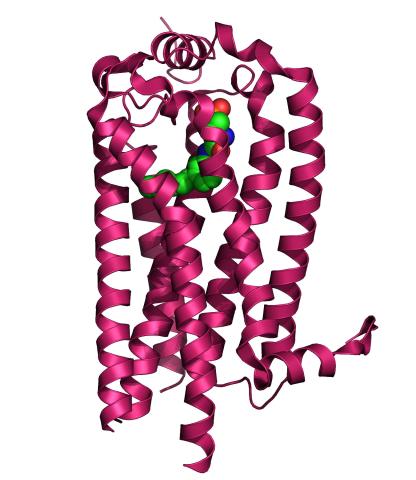
3362: Sphingolipid S1P1 receptor
3362: Sphingolipid S1P1 receptor
The receptor is shown bound to an antagonist, ML056.
Raymond Stevens, The Scripps Research Institute
View Media
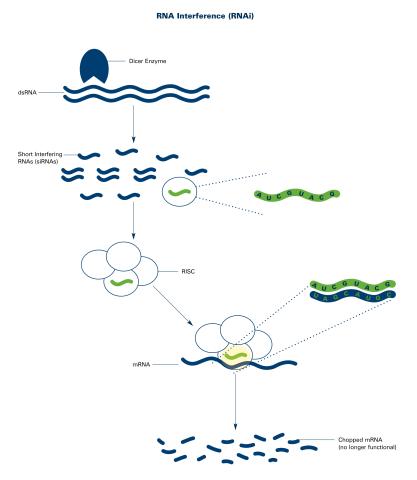
2559: RNA interference (with labels)
2559: RNA interference (with labels)
RNA interference or RNAi is a gene-silencing process in which double-stranded RNAs trigger the destruction of specific RNAs. See 2558 for an unlabeled version of this illustration. Featured in The New Genetics.
Crabtree + Company
View Media
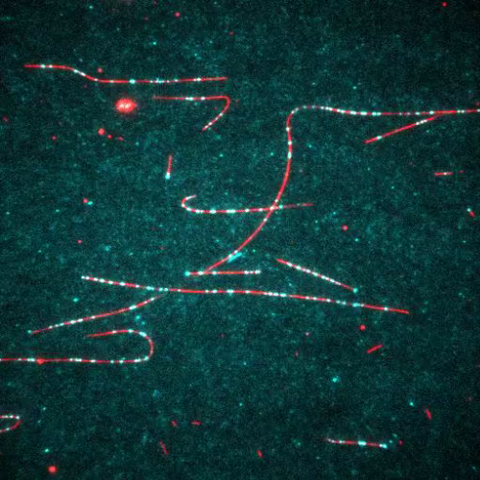
7023: Dynein moving along microtubules
7023: Dynein moving along microtubules
Dynein (green) is a motor protein that “walks” along microtubules (red, part of the cytoskeleton) and carries its cargo along with it. This video was captured through fluorescence microscopy.
Morgan DeSantis, University of Michigan.
View Media
5811: NCMIR Tongue 2
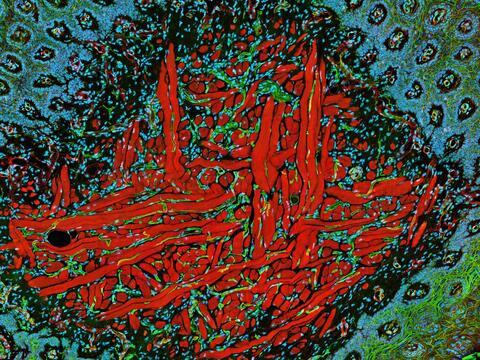
5811: NCMIR Tongue 2
Microscopy image of a tongue. One in a series of two, see image 5810
National Center for Microscopy and Imaging Research (NCMIR)
View Media

6550: Time-lapse video of floral pattern in a mixture of two bacterial species, Acinetobacter baylyi and Escherichia coli, grown on a semi-solid agar for 24 hours
6550: Time-lapse video of floral pattern in a mixture of two bacterial species, Acinetobacter baylyi and Escherichia coli, grown on a semi-solid agar for 24 hours
This time-lapse video shows the emergence of a flower-like pattern in a mixture of two bacterial species, motile Acinetobacter baylyi and non-motile Escherichia coli (green), that are grown together for 24 hours on 0.75% agar surface from a small inoculum in the center of a Petri dish.
See 6557 for a photo of this process at 24 hours on 0.75% agar surface.
See 6553 for a photo of this process at 48 hours on 1% agar surface.
See 6555 for another photo of this process at 48 hours on 1% agar surface.
See 6556 for a photo of this process at 72 hours on 0.5% agar surface.
See 6557 for a photo of this process at 24 hours on 0.75% agar surface.
See 6553 for a photo of this process at 48 hours on 1% agar surface.
See 6555 for another photo of this process at 48 hours on 1% agar surface.
See 6556 for a photo of this process at 72 hours on 0.5% agar surface.
L. Xiong et al, eLife 2020;9: e48885
View Media

6355: H1N1 Influenza Virus
6355: H1N1 Influenza Virus
CellPack image of the H1N1 influenza virus, with hemagglutinin and neuraminidase glycoproteins in green and red, respectively, on the outer envelope (white); matrix protein in gray, and ribonucleoprotein particles inside the virus in red and green. Related to image 6356.
Dr. Rommie Amaro, University of California, San Diego
View Media
2333: Worms and human infertility
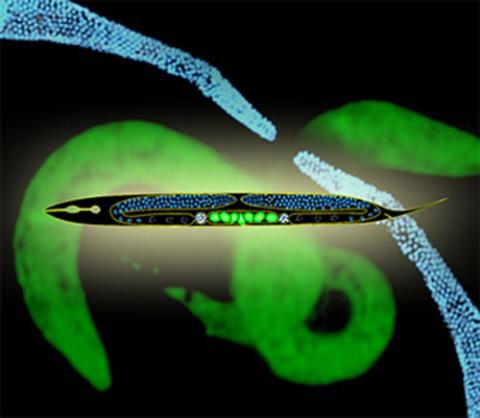
2333: Worms and human infertility
This montage of tiny, transparent C. elegans--or roundworms--may offer insight into understanding human infertility. Researchers used fluorescent dyes to label the worm cells and watch the process of sex cell division, called meiosis, unfold as nuclei (blue) move through the tube-like gonads. Such visualization helps the scientists identify mechanisms that enable these roundworms to reproduce successfully. Because meiosis is similar in all sexually reproducing organisms, what the scientists learn could apply to humans.
Abby Dernburg, Lawrence Berkeley National Laboratory
View Media

1271: Cone cell
1271: Cone cell
The cone cell of the eye allows you to see in color. Appears in the NIGMS booklet Inside the Cell.
Judith Stoffer
View Media
3500: Wound healing in process
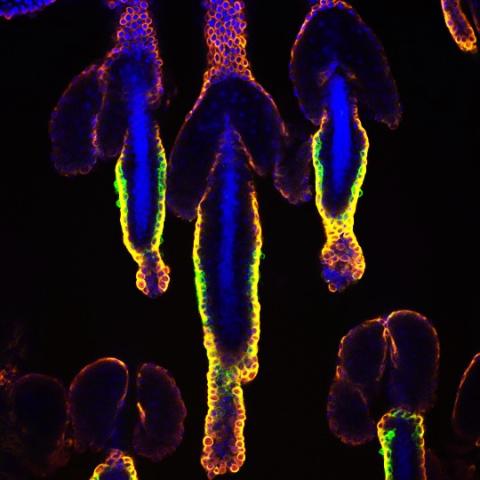
3500: Wound healing in process
Wound healing requires the action of stem cells. In mice that lack the Sept2/ARTS gene, stem cells involved in wound healing live longer and wounds heal faster and more thoroughly than in normal mice. This confocal microscopy image from a mouse lacking the Sept2/ARTS gene shows a tail wound in the process of healing. See more information in the article in Science.
Related to images 3497 and 3498.
Related to images 3497 and 3498.
Hermann Steller, Rockefeller University
View Media
1278: Golgi theories
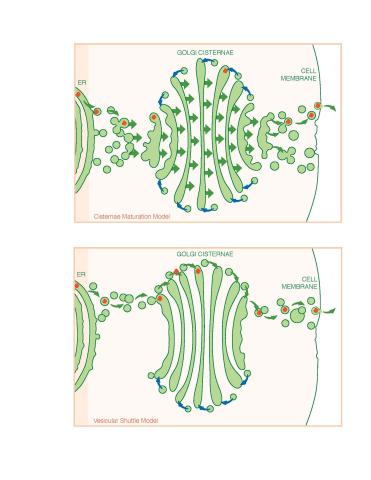
1278: Golgi theories
Two models for how material passes through the Golgi apparatus: the vesicular shuttle model and the cisternae maturation model.
Judith Stoffer
View Media
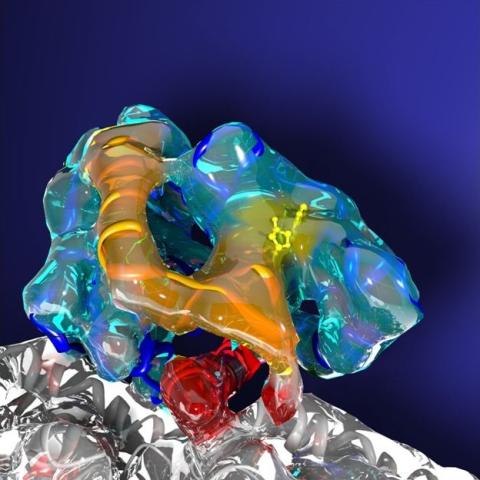
3491: Kinesin moves cellular cargo
3491: Kinesin moves cellular cargo
A protein called kinesin (blue) is in charge of moving cargo around inside cells and helping them divide. It's powered by biological fuel called ATP (bright yellow) as it scoots along tube-like cellular tracks called microtubules (gray).
Charles Sindelar, Yale University
View Media
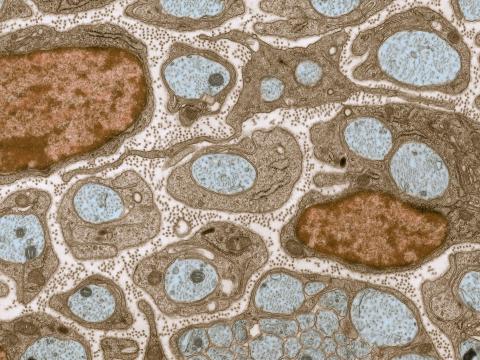
3736: Transmission electron microscopy of myelinated axons with ECM between the axons
3736: Transmission electron microscopy of myelinated axons with ECM between the axons
The extracellular matrix (ECM) is most prevalent in connective tissues but also is present between the stems (axons) of nerve cells, as shown here. Blue-colored nerve cell axons are surrounded by brown-colored, myelin-supplying Schwann cells, which act like insulation around an electrical wire to help speed the transmission of electric nerve impulses down the axon. The ECM is pale pink. The tiny brown spots within it are the collagen fibers that are part of the ECM.
Tom Deerinck, National Center for Microscopy and Imaging Research (NCMIR)
View Media

7014: Flagellated bacterial cells
7014: Flagellated bacterial cells
Vibrio fischeri (2 mm in length) is the exclusive symbiotic partner of the Hawaiian bobtail squid, Euprymna scolopes. After this bacterium uses its flagella to swim from the seawater into the light organ of a newly hatched juvenile, it colonizes the host and loses the appendages. This image was taken using a scanning electron microscope.
Margaret J. McFall-Ngai, Carnegie Institution for Science/California Institute of Technology, and Edward G. Ruby, California Institute of Technology.
View Media
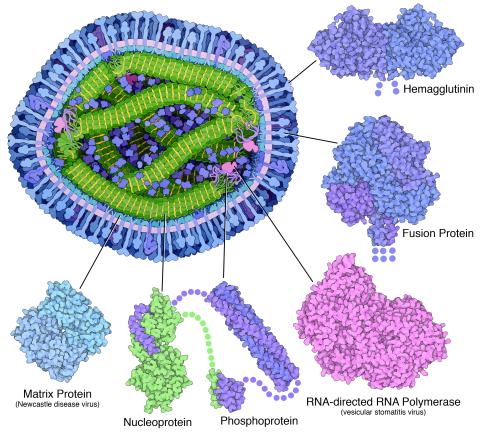
6996: Measles virus proteins
6996: Measles virus proteins
A cross section of the measles virus in which six proteins (enlarged on the outside of the virus) work together to infect cells. The measles virus is extremely infectious; 9 out of 10 people exposed will contract the disease. Fortunately, an effective vaccine protects against infection. Portions of the proteins that have not been determined are shown with dots.
Learn more about the six proteins on PDB 101’s Molecule of the Month: Measles Virus Proteins. Structures are available for the ordered regions of nucleoprotein and phosphoprotein (PDB entries 5E4V, 3ZDO, 1T6O), but the remaining regions are thought to form a flexible, random tangle. For a larger look at the measles virus, see 6995.
Learn more about the six proteins on PDB 101’s Molecule of the Month: Measles Virus Proteins. Structures are available for the ordered regions of nucleoprotein and phosphoprotein (PDB entries 5E4V, 3ZDO, 1T6O), but the remaining regions are thought to form a flexible, random tangle. For a larger look at the measles virus, see 6995.
Amy Wu and Christine Zardecki, RCSB Protein Data Bank.
View Media
6776: Tracking cells in a gastrulating zebrafish embryo
6776: Tracking cells in a gastrulating zebrafish embryo
During development, a zebrafish embryo is transformed from a ball of cells into a recognizable body plan by sweeping convergence and extension cell movements. This process is called gastrulation. Each line in this video represents the movement of a single zebrafish embryo cell over the course of 3 hours. The video was created using time-lapse confocal microscopy. Related to image 6775.
Liliana Solnica-Krezel, Washington University School of Medicine in St. Louis.
View Media