Switch to List View
Image and Video Gallery
This is a searchable collection of scientific photos, illustrations, and videos. The images and videos in this gallery are licensed under Creative Commons Attribution Non-Commercial ShareAlike 3.0. This license lets you remix, tweak, and build upon this work non-commercially, as long as you credit and license your new creations under identical terms.
1081: Natcher Building 01
1081: Natcher Building 01
NIGMS staff are located in the Natcher Building on the NIH campus.
Alisa Machalek, National Institute of General Medical Sciences
View Media
2791: Anti-tumor drug ecteinascidin 743 (ET-743) with hydrogens 02
2791: Anti-tumor drug ecteinascidin 743 (ET-743) with hydrogens 02
Ecteinascidin 743 (ET-743, brand name Yondelis), was discovered and isolated from a sea squirt, Ecteinascidia turbinata, by NIGMS grantee Kenneth Rinehart at the University of Illinois. It was synthesized by NIGMS grantees E.J. Corey and later by Samuel Danishefsky. Multiple versions of this structure are available as entries 2790-2797.
Timothy Jamison, Massachusetts Institute of Technology
View Media
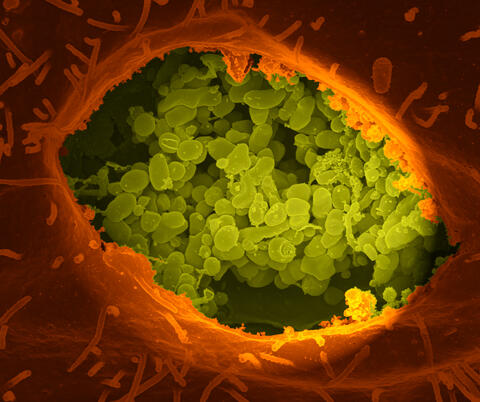
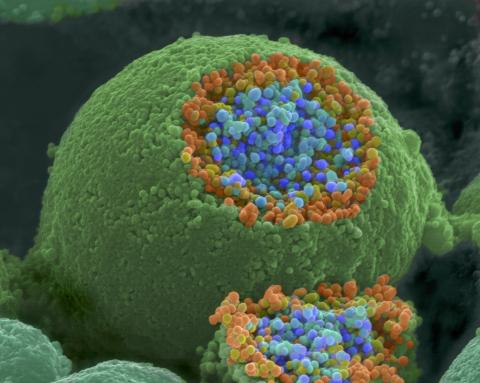
3621: Q fever bacteria in an infected cell
3621: Q fever bacteria in an infected cell
This image shows Q fever bacteria (yellow), which infect cows, sheep, and goats around the world and can infect humans, as well. When caught early, Q fever can be cured with antibiotics. A small fraction of people can develop a more serious, chronic form of the disease.
This image was part of the Life: Magnified exhibit that ran from June 3, 2014, to January 21, 2015, at Dulles International Airport.
This image was part of the Life: Magnified exhibit that ran from June 3, 2014, to January 21, 2015, at Dulles International Airport.
Robert Heinzen, Elizabeth Fischer, and Anita Mora, National Institute of Allergy and Infectious Diseases, National Institutes of Health
View Media
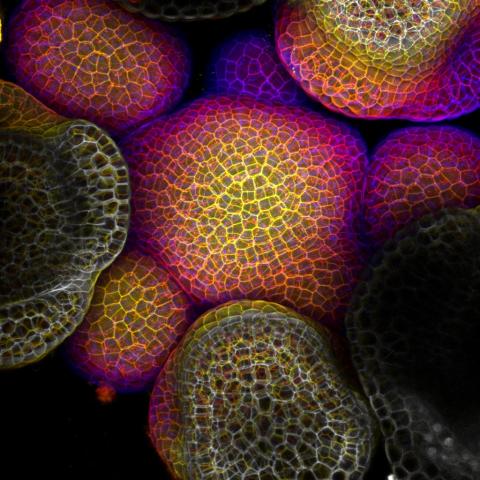
3606: Flower-forming cells in a small plant related to cabbage (Arabidopsis)
3606: Flower-forming cells in a small plant related to cabbage (Arabidopsis)
In plants, as in animals, stem cells can transform into a variety of different cell types. The stem cells at the growing tip of this Arabidopsis plant will soon become flowers. Arabidopsis is frequently studied by cellular and molecular biologists because it grows rapidly (its entire life cycle is only 6 weeks), produces lots of seeds, and has a genome that is easy to manipulate.
This image was part of the Life: Magnified exhibit that ran from June 3, 2014, to January 21, 2015, at Dulles International Airport.
This image was part of the Life: Magnified exhibit that ran from June 3, 2014, to January 21, 2015, at Dulles International Airport.
Arun Sampathkumar and Elliot Meyerowitz, California Institute of Technology
View Media
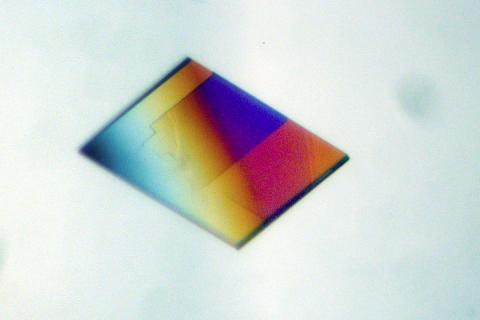
2401: Bacterial alpha amylase
2401: Bacterial alpha amylase
A crystal of bacterial alpha amylase protein created for X-ray crystallography, which can reveal detailed, three-dimensional protein structures.
Alex McPherson, University of California, Irvine
View Media
6597: Pathways – Bacteria vs. Viruses: What's the Difference?
6597: Pathways – Bacteria vs. Viruses: What's the Difference?
Learn about how bacteria and viruses differ, how they each can make you sick, and how they can or cannot be treated. Discover more resources from NIGMS’ Pathways collaboration with Scholastic. View the video on YouTube for closed captioning.
National Institute of General Medical Sciences
View Media
1086: Natcher Building 06
1086: Natcher Building 06
NIGMS staff are located in the Natcher Building on the NIH campus.
Alisa Machalek, National Institute of General Medical Sciences
View Media
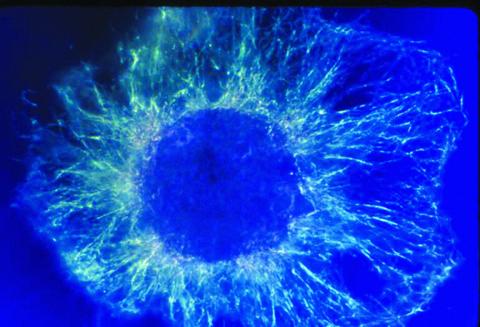
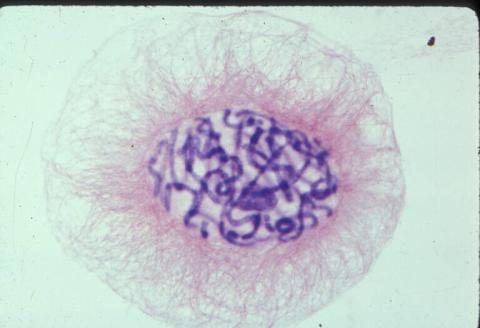
1058: Lily mitosis 01
1058: Lily mitosis 01
A light microscope image shows the chromosomes, stained dark blue, in a dividing cell of an African globe lily (Scadoxus katherinae). This is one frame of a time-lapse sequence that shows cell division in action. The lily is considered a good organism for studying cell division because its chromosomes are much thicker and easier to see than human ones.
Andrew S. Bajer, University of Oregon, Eugene
View Media
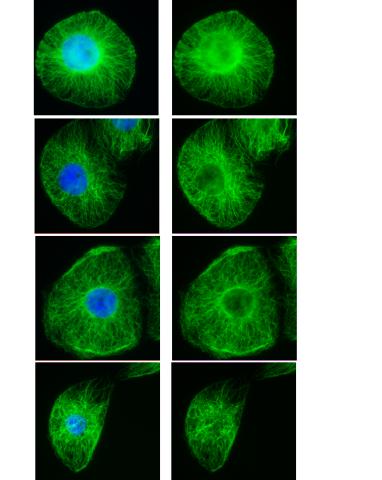
3443: Interphase in Xenopus frog cells
3443: Interphase in Xenopus frog cells
These images show frog cells in interphase. The cells are Xenopus XL177 cells, which are derived from tadpole epithelial cells. The microtubules are green and the chromosomes are blue. Related to 3442.
Claire Walczak, who took them while working as a postdoc in the laboratory of Timothy Mitchison.
View Media
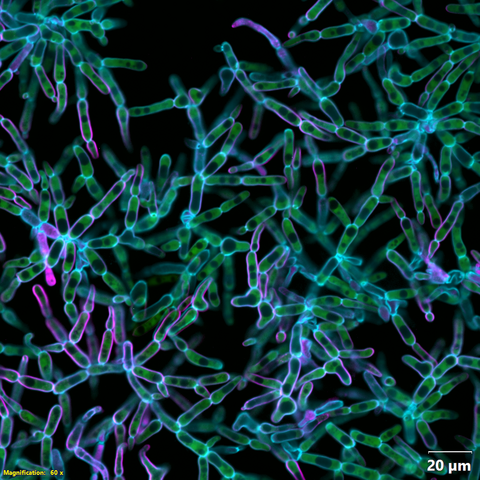
6970: Snowflake yeast 2
6970: Snowflake yeast 2
Multicellular yeast called snowflake yeast that researchers created through many generations of directed evolution from unicellular yeast. Cells are connected to one another by their cell walls, shown in blue. Stained cytoplasm (green) and membranes (magenta) show that the individual cells remain separate. This image was captured using spinning disk confocal microscopy.
Related to images 6969 and 6971.
Related to images 6969 and 6971.
William Ratcliff, Georgia Institute of Technology.
View Media
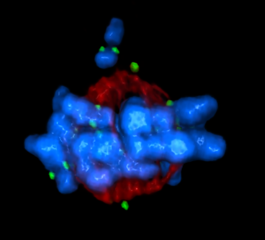
5765: Mitotic cell awaits chromosome alignment
5765: Mitotic cell awaits chromosome alignment
During mitosis, spindle microtubules (red) attach to chromosome pairs (blue), directing them to the spindle equator. This midline alignment is critical for equal distribution of chromosomes in the dividing cell. Scientists are interested in how the protein kinase Plk1 (green) regulates this activity in human cells. Image is a volume projection of multiple deconvolved z-planes acquired with a Nikon widefield fluorescence microscope. This image was chosen as a winner of the 2016 NIH-funded research image call. Related to image 5766.
The research that led to this image was funded by NIGMS.
View Media
The research that led to this image was funded by NIGMS.
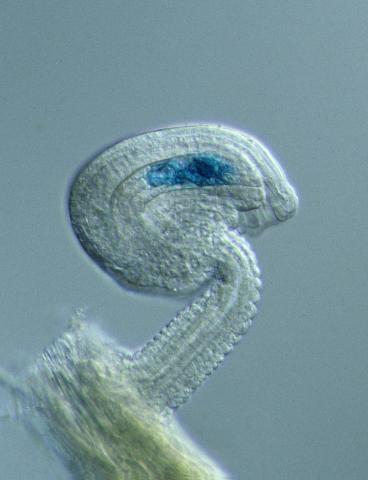
2418: Genetic imprinting in Arabidopsis
2418: Genetic imprinting in Arabidopsis
This delicate, birdlike projection is an immature seed of the Arabidopsis plant. The part in blue shows the cell that gives rise to the endosperm, the tissue that nourishes the embryo. The cell is expressing only the maternal copy of a gene called MEDEA. This phenomenon, in which the activity of a gene can depend on the parent that contributed it, is called genetic imprinting. In Arabidopsis, the maternal copy of MEDEA makes a protein that keeps the paternal copy silent and reduces the size of the endosperm. In flowering plants and mammals, this sort of genetic imprinting is thought to be a way for the mother to protect herself by limiting the resources she gives to any one embryo. Featured in the May 16, 2006, issue of Biomedical Beat.
Robert Fischer, University of California, Berkeley
View Media
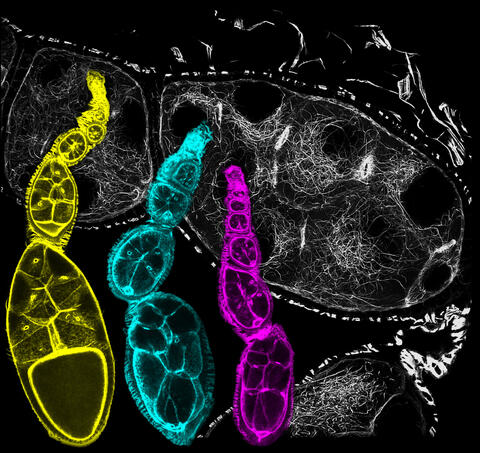
6810: Fruit fly ovarioles
6810: Fruit fly ovarioles
Three fruit fly (Drosophila melanogaster) ovarioles (yellow, blue, and magenta) with egg cells visible inside them. Ovarioles are tubes in the reproductive systems of female insects. Egg cells form at one end of an ovariole and complete their development as they reach the other end, as shown in the yellow wild-type ovariole. This process requires an important protein that is missing in the blue and magenta ovarioles. This image was created using confocal microscopy.
More information on the research that produced this image can be found in the Current Biology paper “Gatekeeper function for Short stop at the ring canals of the Drosophila ovary” by Lu et al.
More information on the research that produced this image can be found in the Current Biology paper “Gatekeeper function for Short stop at the ring canals of the Drosophila ovary” by Lu et al.
Vladimir I. Gelfand, Feinberg School of Medicine, Northwestern University.
View Media
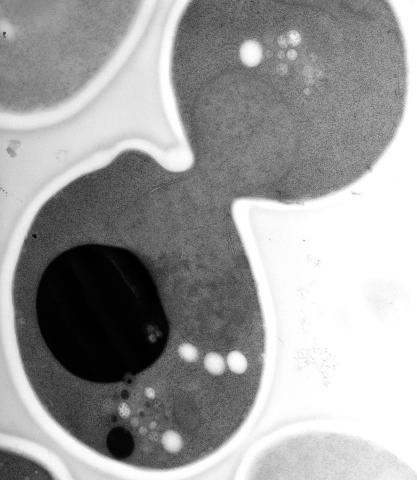
5770: EM of yeast cell division
5770: EM of yeast cell division
Cell division is an incredibly coordinated process. It not only ensures that the new cells formed during this event have a full set of chromosomes, but also that they are endowed with all the cellular materials, including proteins, lipids and small functional compartments called organelles, that are required for normal cell activity. This proper apportioning of essential cell ingredients helps each cell get off to a running start.
This image shows an electron microscopy (EM) thin section taken at 10,000x magnification of a dividing yeast cell over-expressing the protein ubiquitin, which is involved in protein degradation and recycling. The picture features mother and daughter endosome accumulations (small organelles with internal vesicles), a darkly stained vacuole and a dividing nucleus in close contact with a cadre of lipid droplets (unstained spherical bodies). Other dynamic events are also visible, such as spindle microtubules in the nucleus and endocytic pits at the plasma membrane.
These extensive details were revealed thanks to a preservation method involving high-pressure freezing, freeze-substitution and Lowicryl HM20 embedding.
This image shows an electron microscopy (EM) thin section taken at 10,000x magnification of a dividing yeast cell over-expressing the protein ubiquitin, which is involved in protein degradation and recycling. The picture features mother and daughter endosome accumulations (small organelles with internal vesicles), a darkly stained vacuole and a dividing nucleus in close contact with a cadre of lipid droplets (unstained spherical bodies). Other dynamic events are also visible, such as spindle microtubules in the nucleus and endocytic pits at the plasma membrane.
These extensive details were revealed thanks to a preservation method involving high-pressure freezing, freeze-substitution and Lowicryl HM20 embedding.
Matthew West and Greg Odorizzi, University of Colorado
View Media
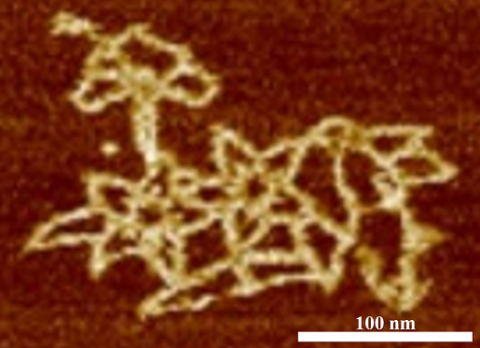
3690: Microscopy image of bird-and-flower DNA origami
3690: Microscopy image of bird-and-flower DNA origami
An atomic force microscopy image shows DNA folded into an intricate, computer-designed structure. Image is featured on Biomedical Beat blog post Cool Image: DNA Origami. See also related image 3689 .
Hao Yan, Arizona State University
View Media
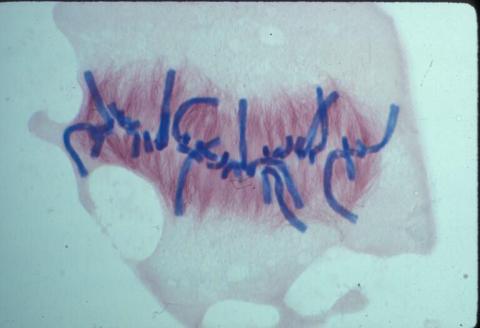
1021: Lily mitosis 08
1021: Lily mitosis 08
A light microscope image of a cell from the endosperm of an African globe lily (Scadoxus katherinae). This is one frame of a time-lapse sequence that shows cell division in action. The lily is considered a good organism for studying cell division because its chromosomes are much thicker and easier to see than human ones. Staining shows microtubules in red and chromosomes in blue. Here, condensed chromosomes are clearly visible and lined up.
Related to images 1010, 1011, 1012, 1013, 1014, 1015, 1016, 1017, 1018, and 1019.
Related to images 1010, 1011, 1012, 1013, 1014, 1015, 1016, 1017, 1018, and 1019.
Andrew S. Bajer, University of Oregon, Eugene
View Media

2385: Heat shock protein complex from Methanococcus jannaschii
2385: Heat shock protein complex from Methanococcus jannaschii
Model based on X-ray crystallography of the structure of a small heat shock protein complex from the bacteria, Methanococcus jannaschii. Methanococcus jannaschii is an organism that lives at near boiling temperature, and this protein complex helps it cope with the stress of high temperature. Similar complexes are produced in human cells when they are "stressed" by events such as burns, heart attacks, or strokes. The complexes help cells recover from the stressful event.
Berkeley Structural Genomics Center, PSI-1
View Media
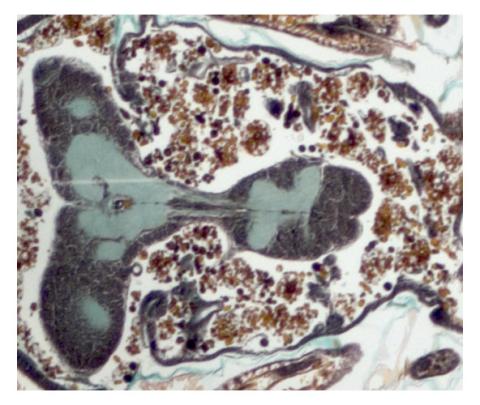
2758: Cross section of a Drosophila melanogaster pupa
2758: Cross section of a Drosophila melanogaster pupa
This photograph shows a magnified view of a Drosophila melanogaster pupa in cross section. Compare this normal pupa to one that lacks an important receptor, shown in image 2759.
Christina McPhee and Eric Baehrecke, University of Massachusetts Medical School
View Media
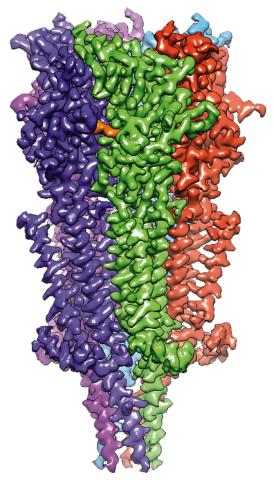
6579: Full-length serotonin receptor (ion channel)
6579: Full-length serotonin receptor (ion channel)
A 3D reconstruction, created using cryo-electron microscopy, of an ion channel known as the full-length serotonin receptor in complex with the antinausea drug granisetron (orange). Ion channels are proteins in cell membranes that help regulate many processes.
Sudha Chakrapani, Case Western Reserve University School of Medicine.
View Media
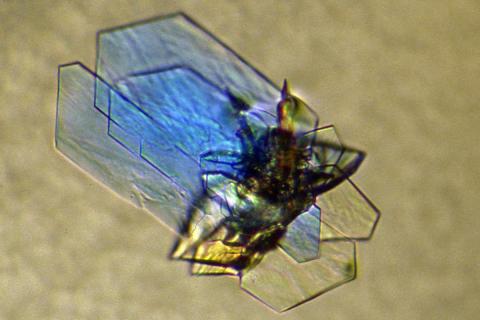
2410: DNase
2410: DNase
Crystals of DNase protein created for X-ray crystallography, which can reveal detailed, three-dimensional protein structures.
Alex McPherson, University of California, Irvine
View Media

6771: Culex quinquefasciatus mosquito larvae
6771: Culex quinquefasciatus mosquito larvae
Mosquito larvae with genes edited by CRISPR swimming in water. This species of mosquito, Culex quinquefasciatus, can transmit West Nile virus, Japanese encephalitis virus, and avian malaria, among other diseases. The researchers who took this video optimized the gene-editing tool CRISPR for Culex quinquefasciatus that could ultimately help stop the mosquitoes from spreading pathogens. The work is described in the Nature Communications paper "Optimized CRISPR tools and site-directed transgenesis towards gene drive development in Culex quinquefasciatus mosquitoes" by Feng et al. Related to images 6769 and 6770.
Valentino Gantz, University of California, San Diego.
View Media
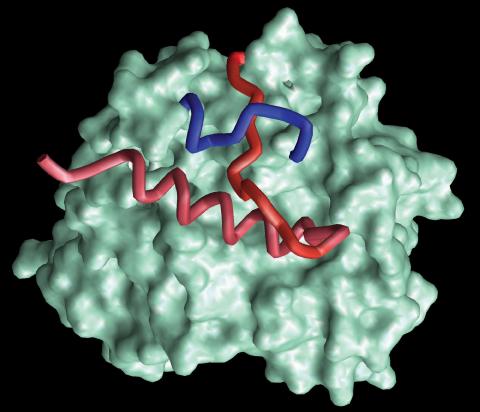
2374: Protein from Methanobacterium thermoautotrophicam
2374: Protein from Methanobacterium thermoautotrophicam
A knotted protein from an archaebacterium called Methanobacterium thermoautotrophicam. This organism breaks down waste products and produces methane gas. Protein folding theory previously held that forming a knot was beyond the ability of a protein, but this structure, determined at Argonne's Structural Biology Center, proves differently. Researchers theorize that this knot stabilizes the amino acid subunits of the protein.
Midwest Center For Structural Genomics, PSI
View Media
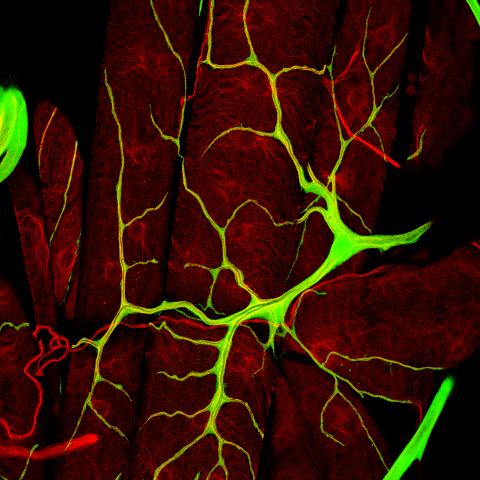
3615: An insect tracheal cell delivers air to muscles
3615: An insect tracheal cell delivers air to muscles
Insects like the fruit fly use an elaborate network of branching tubes called trachea (green) to transport oxygen throughout their bodies. Fruit flies have been used in biomedical research for more than 100 years and remain one of the most frequently studied model organisms. They have a large percentage of genes in common with us, including hundreds of genes that are associated with human diseases.
This image was part of the Life: Magnified exhibit that ran from June 3, 2014, to January 21, 2015, at Dulles International Airport.
This image was part of the Life: Magnified exhibit that ran from June 3, 2014, to January 21, 2015, at Dulles International Airport.
Jayan Nair and Maria Leptin, European Molecular Biology Laboratory, Heidelberg, Germany
View Media
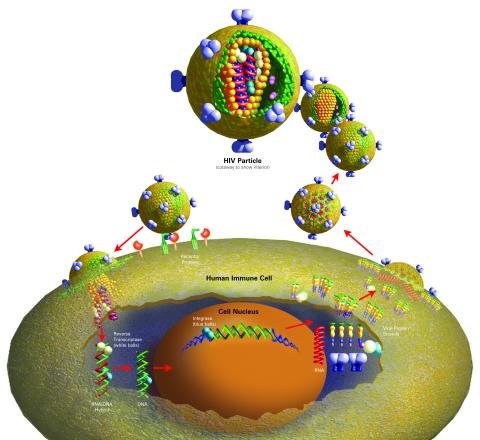
2514: Life of an AIDS virus (with labels)
2514: Life of an AIDS virus (with labels)
HIV is a retrovirus, a type of virus that carries its genetic material not as DNA but as RNA. Long before anyone had heard of HIV, researchers in labs all over the world studied retroviruses, tracing out their life cycle and identifying the key proteins the viruses use to infect cells. When HIV was identified as a retrovirus, these studies gave AIDS researchers an immediate jump-start. The previously identified viral proteins became initial drug targets. See images 2513 and 2515 for other versions of this illustration. Featured in The Structures of Life.
Crabtree + Company
View Media
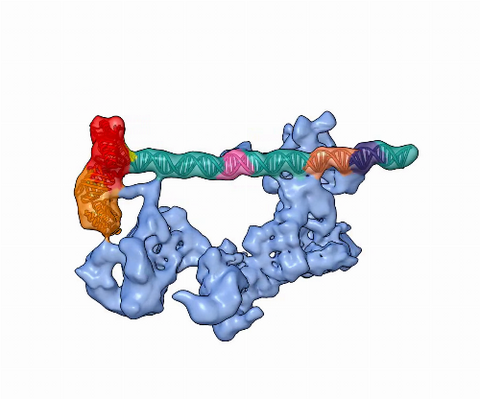
5730: Dynamic cryo-EM model of the human transcription preinitiation complex
5730: Dynamic cryo-EM model of the human transcription preinitiation complex
Gene transcription is a process by which information encoded in DNA is transcribed into RNA. It's essential for all life and requires the activity of proteins, called transcription factors, that detect where in a DNA strand transcription should start. In eukaryotes (i.e., those that have a nucleus and mitochondria), a protein complex comprising 14 different proteins is responsible for sniffing out transcription start sites and starting the process. This complex represents the core machinery to which an enzyme, named RNA polymerase, can bind to and read the DNA and transcribe it to RNA. Scientists have used cryo-electron microscopy (cryo-EM) to visualize the TFIID-RNA polymerase-DNA complex in unprecedented detail. This animation shows the different TFIID components as they contact DNA and recruit the RNA polymerase for gene transcription.
To learn more about the research that has shed new light on gene transcription, see this news release from Berkeley Lab.
Related to image 3766.
To learn more about the research that has shed new light on gene transcription, see this news release from Berkeley Lab.
Related to image 3766.
Eva Nogales, Berkeley Lab
View Media

5755: Autofluorescent xanthophores in zebrafish skin
5755: Autofluorescent xanthophores in zebrafish skin
Pigment cells are cells that give skin its color. In fishes and amphibians, like frogs and salamanders, pigment cells are responsible for the characteristic skin patterns that help these organisms to blend into their surroundings or attract mates. The pigment cells are derived from neural crest cells, which are cells originating from the neural tube in the early embryo. This image shows pigment cells called xanthophores in the skin of zebrafish; the cells glow (autofluoresce) brightly under light giving the fish skin a shiny, lively appearance. Investigating pigment cell formation and migration in animals helps answer important fundamental questions about the factors that control pigmentation in the skin of animals, including humans. Related to images 5754, 5756, 5757 and 5758.
David Parichy, University of Washington
View Media
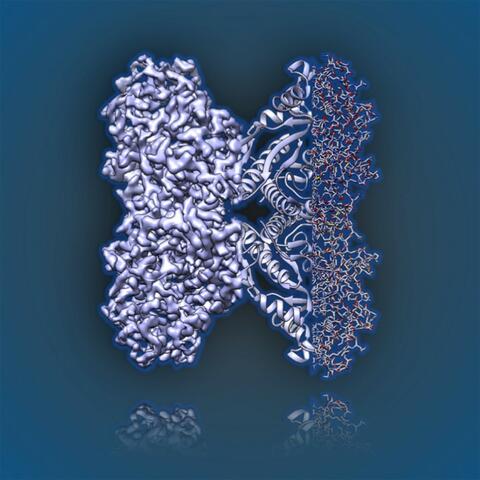
6350: Aldolase
6350: Aldolase
2.5Å resolution reconstruction of rabbit muscle aldolase collected on a FEI/Thermo Fisher Titan Krios with energy filter and image corrector.
National Resource for Automated Molecular Microscopy http://nramm.nysbc.org/nramm-images/ Source: Bridget Carragher
View Media
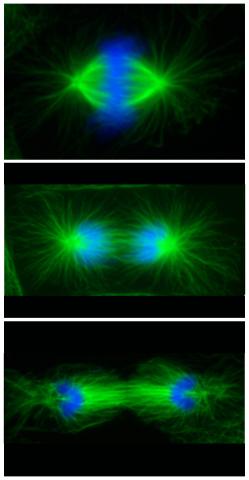
3442: Cell division phases in Xenopus frog cells
3442: Cell division phases in Xenopus frog cells
These images show three stages of cell division in Xenopus XL177 cells, which are derived from tadpole epithelial cells. They are (from top): metaphase, anaphase and telophase. The microtubules are green and the chromosomes are blue. Related to 3443.
Claire Walczak, who took them while working as a postdoc in the laboratory of Timothy Mitchison
View Media
1244: Nerve ending
1244: Nerve ending
A scanning electron microscope picture of a nerve ending. It has been broken open to reveal vesicles (orange and blue) containing chemicals used to pass messages in the nervous system.
Tina Weatherby Carvalho, University of Hawaii at Manoa
View Media
1014: Lily mitosis 04
1014: Lily mitosis 04
A light microscope image of a cell from the endosperm of an African globe lily (Scadoxus katherinae). This is one frame of a time-lapse sequence that shows cell division in action. The lily is considered a good organism for studying cell division because its chromosomes are much thicker and easier to see than human ones. Staining shows microtubules in red and chromosomes in blue.
Related to images 1010, 1011, 1012, 1013, 1015, 1016, 1017, 1018, 1019, and 1021.
Related to images 1010, 1011, 1012, 1013, 1015, 1016, 1017, 1018, 1019, and 1021.
Andrew S. Bajer, University of Oregon, Eugene
View Media

2384: Scientists display X-ray diffraction pattern obtained with split X-ray beamline
2384: Scientists display X-ray diffraction pattern obtained with split X-ray beamline
Scientists from Argonne National Laboratory's Advanced Photon Source (APS) display the first X-ray diffraction pattern obtained from a protein crystal using a split X-ray beam, the first of its kind at APS. The scientists shown are (from left to right): Oleg Makarov, Ruslan Sanishvili, Robert Fischetti (project manager), Sergey Stepanov, and Ward Smith.
GM/CA Collaborative Access Team
View Media
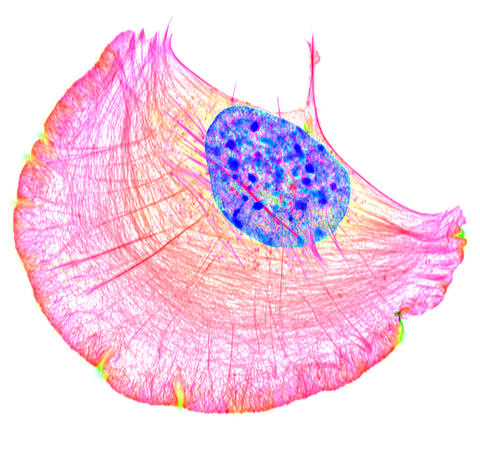
6964: Crawling cell
6964: Crawling cell
A crawling cell with DNA shown in blue and actin filaments, which are a major component of the cytoskeleton, visible in pink. Actin filaments help enable cells to crawl. This image was captured using structured illumination microscopy.
Dylan T. Burnette, Vanderbilt University School of Medicine.
View Media
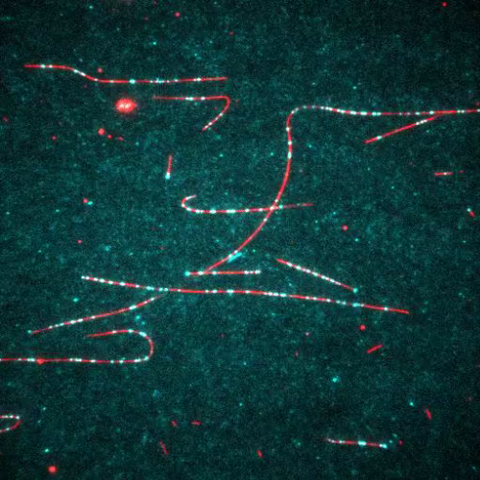
7023: Dynein moving along microtubules
7023: Dynein moving along microtubules
Dynein (green) is a motor protein that “walks” along microtubules (red, part of the cytoskeleton) and carries its cargo along with it. This video was captured through fluorescence microscopy.
Morgan DeSantis, University of Michigan.
View Media
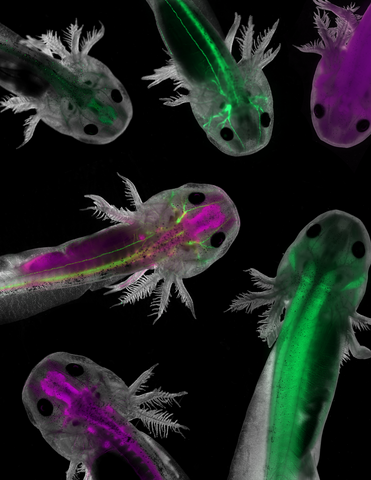
6928: Axolotls showing nervous system components
6928: Axolotls showing nervous system components
Axolotls—a type of salamander—that have been genetically modified so that various parts of their nervous systems glow purple and green. Researchers often study axolotls for their extensive regenerative abilities. They can regrow tails, limbs, spinal cords, brains, and more. The researcher who took this image focuses on the role of the peripheral nervous system during limb regeneration.
This image was captured using a stereo microscope.
Related to images 6927 and 6932.
This image was captured using a stereo microscope.
Related to images 6927 and 6932.
Prayag Murawala, MDI Biological Laboratory and Hannover Medical School.
View Media
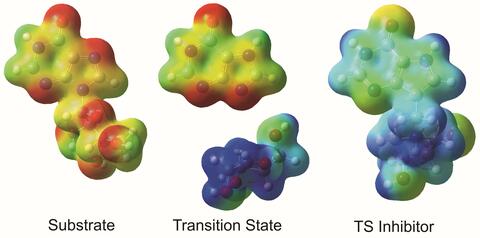
3429: Enzyme transition states
3429: Enzyme transition states
The molecule on the left is an electrostatic potential map of the van der Waals surface of the transition state for human purine nucleoside phosphorylase. The colors indicate the electron density at any position of the molecule. Red indicates electron-rich regions with negative charge and blue indicates electron-poor regions with positive charge. The molecule on the right is called DADMe-ImmH. It is a chemically stable analogue of the transition state on the left. It binds to the enzyme millions of times tighter than the substrate. This inhibitor is in human clinical trials for treating patients with gout. This image appears in Figure 4, Schramm, V.L. (2011) Annu. Rev. Biochem. 80:703-732.
Vern Schramm, Albert Einstein College of Medicine of Yeshiva University
View Media
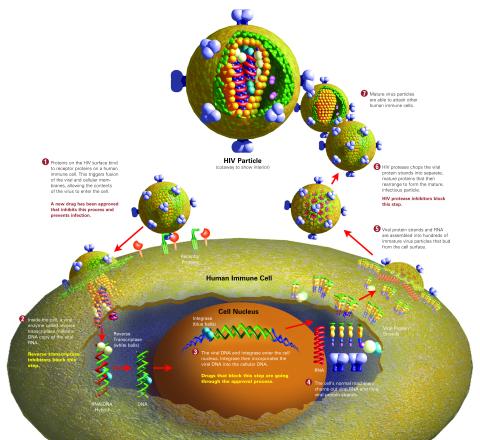
2515: Life of an AIDS virus (with labels and stages)
2515: Life of an AIDS virus (with labels and stages)
HIV is a retrovirus, a type of virus that carries its genetic material not as DNA but as RNA. Long before anyone had heard of HIV, researchers in labs all over the world studied retroviruses, tracing out their life cycle and identifying the key proteins the viruses use to infect cells. When HIV was identified as a retrovirus, these studies gave AIDS researchers an immediate jump-start. The previously identified viral proteins became initial drug targets. See images 2513 and 2514 for other versions of this illustration. Featured in The Structures of Life.
Crabtree + Company
View Media
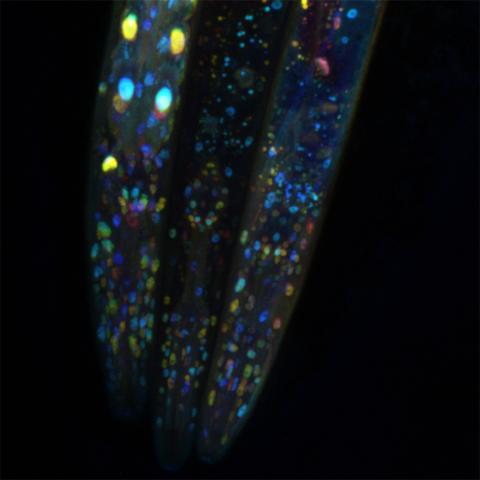
6532: Mosaicism in C. elegans (Black Background)
6532: Mosaicism in C. elegans (Black Background)
In the worm C. elegans, double-stranded RNA made in neurons can silence matching genes in a variety of cell types through the transport of RNA between cells. The head region of three worms that were genetically modified to express a fluorescent protein were imaged and the images were color-coded based on depth. The worm on the left lacks neuronal double-stranded RNA and thus every cell is fluorescent. In the middle worm, the expression of the fluorescent protein is silenced by neuronal double-stranded RNA and thus most cells are not fluorescent. The worm on the right lacks an enzyme that amplifies RNA for silencing. Surprisingly, the identities of the cells that depend on this enzyme for gene silencing are unpredictable. As a result, worms of identical genotype are nevertheless random mosaics for how the function of gene silencing is carried out. For more, see journal article and press release. Related to image 6534.
Snusha Ravikumar, Ph.D., University of Maryland, College Park, and Antony M. Jose, Ph.D., University of Maryland, College Park
View Media
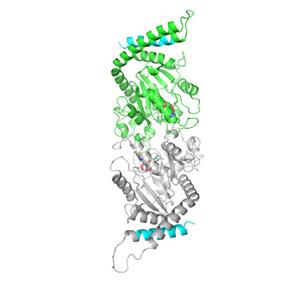
2744: Dynamin structure
2744: Dynamin structure
When a molecule arrives at a cell's outer membrane, the membrane creates a pouch around the molecule that protrudes inward. Directed by a protein called dynamin, the pouch then gets pinched off to form a vesicle that carries the molecule to the right place inside the cell. To better understand how dynamin performs its vital pouch-pinching role, researchers determined its structure. Based on the structure, they proposed that a dynamin "collar" at the pouch's base twists ever tighter until the vesicle pops free. Because cells absorb many drugs through vesicles, the discovery could lead to new drug delivery methods.
Josh Chappie, National Institute of Diabetes and Digestive and Kidney Diseases, NIH
View Media
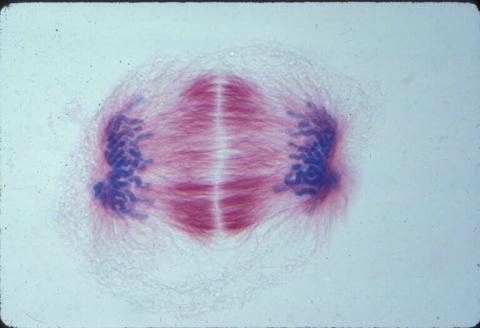
1018: Lily mitosis 12
1018: Lily mitosis 12
A light microscope image of a cell from the endosperm of an African globe lily (Scadoxus katherinae). This is one frame of a time-lapse sequence that shows cell division in action. The lily is considered a good organism for studying cell division because its chromosomes are much thicker and easier to see than human ones. Staining shows microtubules in red and chromosomes in blue. Here, condensed chromosomes are clearly visible near the end of a round of mitosis.
Related to images 1010, 1011, 1012, 1013, 1014, 1015, 1016, 1017, 1019, and 1021.
Related to images 1010, 1011, 1012, 1013, 1014, 1015, 1016, 1017, 1019, and 1021.
Andrew S. Bajer, University of Oregon, Eugene
View Media
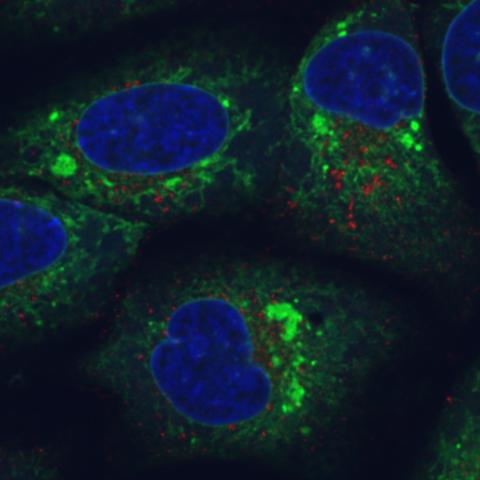
6773: Endoplasmic reticulum abnormalities
6773: Endoplasmic reticulum abnormalities
Human cells with the gene that codes for the protein FIT2 deleted. Green indicates an endoplasmic reticulum (ER) resident protein. The lack of FIT2 affected the structure of the ER and caused the resident protein to cluster in ER membrane aggregates, seen as large, bright-green spots. Red shows where the degradation of cell parts—called autophagy—is taking place, and the nucleus is visible in blue. This image was captured using a confocal microscope.
Michel Becuwe, Harvard University.
View Media

2345: Magnesium transporter protein from E. faecalis
2345: Magnesium transporter protein from E. faecalis
Structure of a magnesium transporter protein from an antibiotic-resistant bacterium (Enterococcus faecalis) found in the human gut. Featured as one of the June 2007 Protein Sructure Initiative Structures of the Month.
New York Structural GenomiX Consortium
View Media
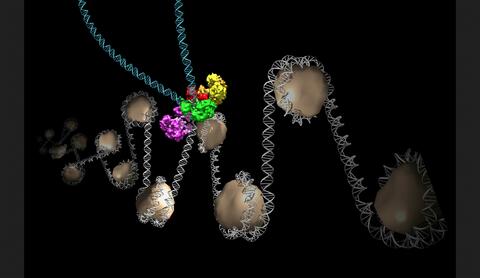
6346: Intasome
6346: Intasome
Salk researchers captured the structure of a protein complex called an intasome (center) that lets viruses similar to HIV establish permanent infection in their hosts. The intasome hijacks host genomic material, DNA (white) and histones (beige), and irreversibly inserts viral DNA (blue). The image was created by Jamie Simon and Dmitry Lyumkis. Work that led to the 3D map was published in: Ballandras-Colas A, Brown M, Cook NJ, Dewdney TG, Demeler B, Cherepanov P, Lyumkis D, & Engelman AN. (2016). Cryo-EM reveals a novel octameric integrase structure for ?-retroviral intasome function. Nature, 530(7590), 358—361
National Resource for Automated Molecular Microscopy http://nramm.nysbc.org/nramm-images/ Source: Bridget Carragher
View Media
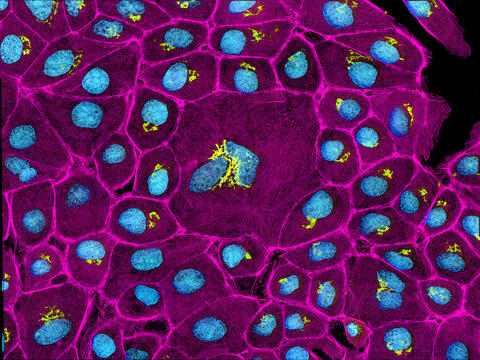
3647: Epithelial cells
3647: Epithelial cells
This image mostly shows normal cultured epithelial cells expressing green fluorescent protein targeted to the Golgi apparatus (yellow-green) and stained for actin (magenta) and DNA (cyan). The middle cell is an abnormal large multinucleated cell. All the cells in this image have a Golgi but not all are expressing the targeted recombinant fluorescent protein.
Tom Deerinck, National Center for Microscopy and Imaging Research (NCMIR)
View Media

2702: Thermotoga maritima and its metabolic network
2702: Thermotoga maritima and its metabolic network
A combination of protein structures determined experimentally and computationally shows us the complete metabolic network of a heat-loving bacterium.
View Media

6550: Time-lapse video of floral pattern in a mixture of two bacterial species, Acinetobacter baylyi and Escherichia coli, grown on a semi-solid agar for 24 hours
6550: Time-lapse video of floral pattern in a mixture of two bacterial species, Acinetobacter baylyi and Escherichia coli, grown on a semi-solid agar for 24 hours
This time-lapse video shows the emergence of a flower-like pattern in a mixture of two bacterial species, motile Acinetobacter baylyi and non-motile Escherichia coli (green), that are grown together for 24 hours on 0.75% agar surface from a small inoculum in the center of a Petri dish.
See 6557 for a photo of this process at 24 hours on 0.75% agar surface.
See 6553 for a photo of this process at 48 hours on 1% agar surface.
See 6555 for another photo of this process at 48 hours on 1% agar surface.
See 6556 for a photo of this process at 72 hours on 0.5% agar surface.
See 6557 for a photo of this process at 24 hours on 0.75% agar surface.
See 6553 for a photo of this process at 48 hours on 1% agar surface.
See 6555 for another photo of this process at 48 hours on 1% agar surface.
See 6556 for a photo of this process at 72 hours on 0.5% agar surface.
L. Xiong et al, eLife 2020;9: e48885
View Media
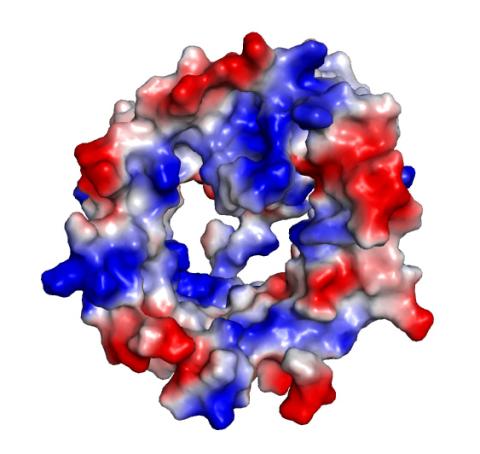
2495: VDAC-1 (4)
2495: VDAC-1 (4)
The structure of the pore-forming protein VDAC-1 from humans. This molecule mediates the flow of products needed for metabolism--in particular the export of ATP--across the outer membrane of mitochondria, the power plants for eukaryotic cells. VDAC-1 is involved in metabolism and the self-destruction of cells--two biological processes central to health.
Related to images 2491, 2494, and 2488.
Related to images 2491, 2494, and 2488.
Gerhard Wagner, Harvard Medical School
View Media
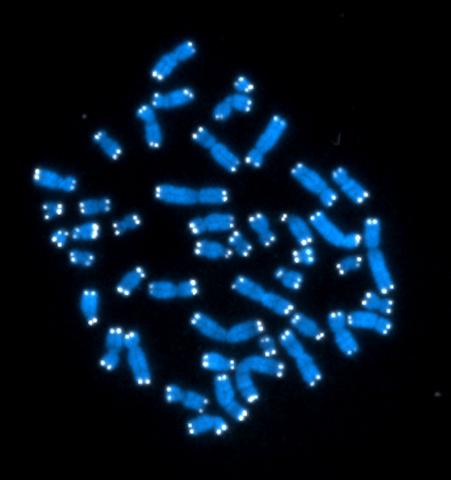
2626: Telomeres
2626: Telomeres
The 46 human chromosomes are shown in blue, with the telomeres appearing as white pinpoints. The DNA has already been copied, so each chromosome is actually made up of two identical lengths of DNA, each with its own two telomeres.
Hesed Padilla-Nash and Thomas Ried, the National Cancer Institute, a part of NIH
View Media
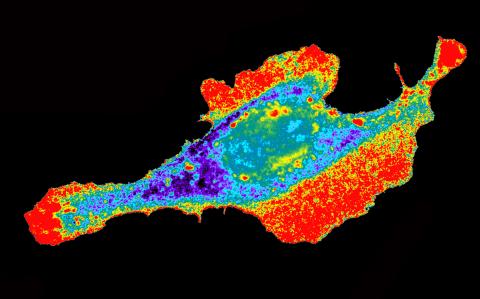
2453: Seeing signaling protein activation in cells 03
2453: Seeing signaling protein activation in cells 03
Cdc42, a member of the Rho family of small guanosine triphosphatase (GTPase) proteins, regulates multiple cell functions, including motility, proliferation, apoptosis, and cell morphology. In order to fulfill these diverse roles, the timing and location of Cdc42 activation must be tightly controlled. Klaus Hahn and his research group use special dyes designed to report protein conformational changes and interactions, here in living neutrophil cells. Warmer colors in this image indicate higher levels of activation. Cdc42 looks to be activated at cell protrusions.
Related to images 2451, 2452, and 2454.
Related to images 2451, 2452, and 2454.
Klaus Hahn, University of North Carolina, Chapel Hill Medical School
View Media
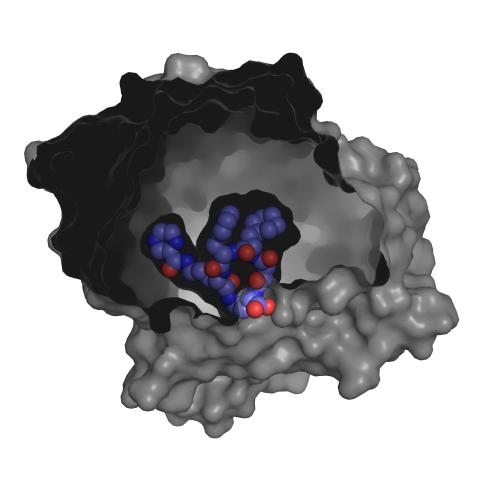
3415: X-ray co-crystal structure of Src kinase bound to a DNA-templated macrocycle inhibitor 3
3415: X-ray co-crystal structure of Src kinase bound to a DNA-templated macrocycle inhibitor 3
X-ray co-crystal structure of Src kinase bound to a DNA-templated macrocycle inhibitor. Related to 3413, 3414, 3416, 3417, 3418, and 3419.
Markus A. Seeliger, Stony Brook University Medical School and David R. Liu, Harvard University
View Media
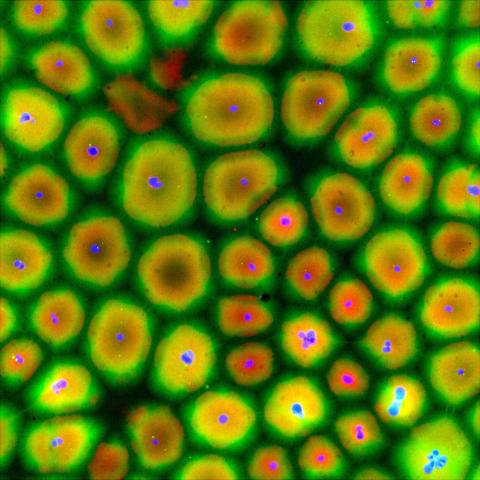
6584: Cell-like compartments from frog eggs
6584: Cell-like compartments from frog eggs
Cell-like compartments that spontaneously emerged from scrambled frog eggs, with nuclei (blue) from frog sperm. Endoplasmic reticulum (red) and microtubules (green) are also visible. Image created using epifluorescence microscopy.
For more photos of cell-like compartments from frog eggs view: 6585, 6586, 6591, 6592, and 6593.
For videos of cell-like compartments from frog eggs view: 6587, 6588, 6589, and 6590.
Xianrui Cheng, Stanford University School of Medicine.
View Media
