Switch to List View
Image and Video Gallery
This is a searchable collection of scientific photos, illustrations, and videos. The images and videos in this gallery are licensed under Creative Commons Attribution Non-Commercial ShareAlike 3.0. This license lets you remix, tweak, and build upon this work non-commercially, as long as you credit and license your new creations under identical terms.
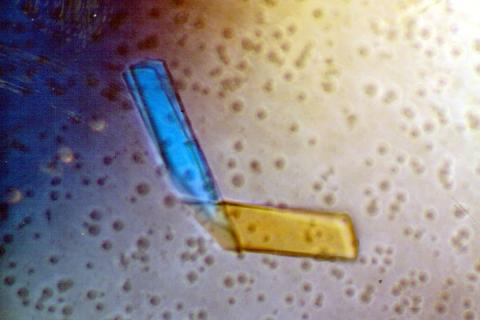
2399: Bence Jones protein MLE
2399: Bence Jones protein MLE
A crystal of Bence Jones protein created for X-ray crystallography, which can reveal detailed, three-dimensional protein structures.
Alex McPherson, University of California, Irvine
View Media
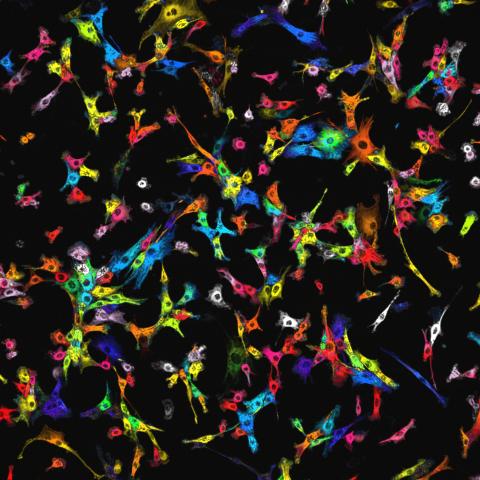
7021: Single-cell “radios” image
7021: Single-cell “radios” image
Individual cells are color-coded based on their identity and signaling activity using a protein circuit technology developed by the Coyle Lab. Just as a radio allows you to listen to an individual frequency, this technology allows researchers to tune into the specific “radio station” of each cell through genetically encoded proteins from a bacterial system called MinDE. The proteins generate an oscillating fluorescent signal that transmits information about cell shape, state, and identity that can be decoded using digital signal processing tools originally designed for telecommunications. The approach allows researchers to look at the dynamics of a single cell in the presence of many other cells.
Related to video 7022.
Related to video 7022.
Scott Coyle, University of Wisconsin-Madison.
View Media
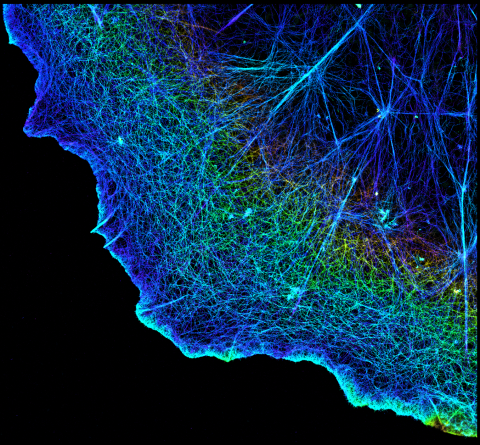
3749: 3D image of actin in a cell
3749: 3D image of actin in a cell
Actin is an essential protein in a cell's skeleton (cytoskeleton). It forms a dense network of thin filaments in the cell. Here, researchers have used a technique called stochastic optical reconstruction microscopy (STORM) to visualize the actin network in a cell in three dimensions. The actin strands were labeled with a dye called Alexa Fluor 647-phalloidin. This image appears in a study published by Nature Methods, which reports how researchers use STORM to visualize the cytoskeleton.
Xiaowei Zhuang, Howard Hughes Medical Institute, Harvard University
View Media
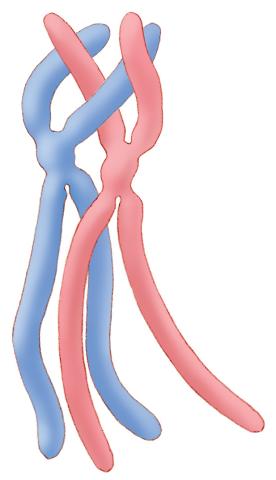
1315: Chromosomes before crossing over
1315: Chromosomes before crossing over
Duplicated pair of chromosomes lined up and ready to cross over.
Judith Stoffer
View Media
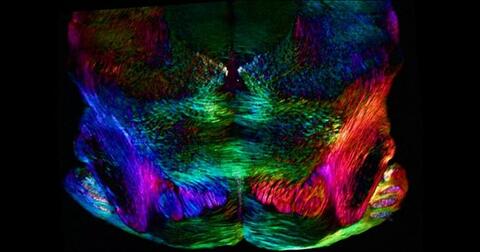
6901: Mouse brain slice showing nerve cells
6901: Mouse brain slice showing nerve cells
A 20-µm thick section of mouse midbrain. The nerve cells are transparent and weren’t stained. Instead, the color is generated by interaction of white polarized light with the molecules in the cells and indicates their orientation.
The image was obtained with a polychromatic polarizing microscope that shows the polychromatic birefringent image with hue corresponding to the slow axis orientation. More information about the microscopy that produced this image can be found in the Scientific Reports paper “Polychromatic Polarization Microscope: Bringing Colors to a Colorless World” by Shribak.
The image was obtained with a polychromatic polarizing microscope that shows the polychromatic birefringent image with hue corresponding to the slow axis orientation. More information about the microscopy that produced this image can be found in the Scientific Reports paper “Polychromatic Polarization Microscope: Bringing Colors to a Colorless World” by Shribak.
Michael Shribak, Marine Biological Laboratory/University of Chicago.
View Media
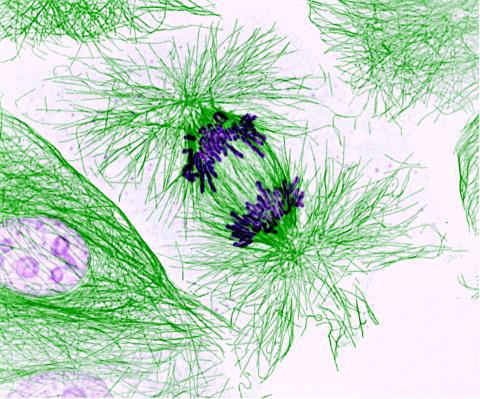
3631: Dividing cells showing chromosomes and cell skeleton
3631: Dividing cells showing chromosomes and cell skeleton
This pig cell is in the process of dividing. The chromosomes (purple) have already replicated and the duplicates are being pulled apart by fibers of the cell skeleton known as microtubules (green). Studies of cell division yield knowledge that is critical to advancing understanding of many human diseases, including cancer and birth defects.
This image was part of the Life: Magnified exhibit that ran from June 3, 2014, to January 21, 2015, at Dulles International Airport.
This image was part of the Life: Magnified exhibit that ran from June 3, 2014, to January 21, 2015, at Dulles International Airport.
Nasser Rusan, National Heart, Lung, and Blood Institute, National Institutes of Health
View Media
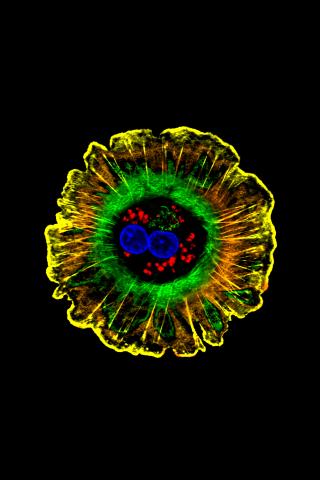
3610: Human liver cell (hepatocyte)
3610: Human liver cell (hepatocyte)
Hepatocytes, like the one shown here, are the most abundant type of cell in the human liver. They play an important role in building proteins; producing bile, a liquid that aids in digesting fats; and chemically processing molecules found normally in the body, like hormones, as well as foreign substances like medicines and alcohol.
This image was part of the Life: Magnified exhibit that ran from June 3, 2014, to January 21, 2015, at Dulles International Airport.
This image was part of the Life: Magnified exhibit that ran from June 3, 2014, to January 21, 2015, at Dulles International Airport.
Donna Beer Stolz, University of Pittsburgh
View Media

5753: Clathrin-mediated endocytosis
5753: Clathrin-mediated endocytosis
Endocytosis is the process by which cells are able to take up membrane and extracellular materials through the formation of a small intracellular bubble, called a vesicle. This process, called membrane budding, is generally by a coating of proteins. This protein coat helps both to deform the membrane and to concentrate specific proteins inside the newly forming vesicle. Clathrin is a coat protein that functions in receptor-mediated endocytosis events at the plasma membrane. This animation shows the process of clathrin-mediated endocytosis. An iron-transport protein called transferrin (blue) is bound to its receptor (purple) on the exterior cell membrane. Inside the cell, a clathrin cage (shown in white/beige) assembles through interactions with membrane-bound adaptor proteins (green), causing the cell membrane to begin bending. The adaptor proteins also bind to receptors for transferrin, capturing them in the growing vesicle. Molecules of a protein called dynamin (purple) are then recruited to the neck of the vesicle and are involved in separating the membranes of the cell and the vesicle. Soon after the vesicle has budded off the membrane, the clathrin cage is disassembled. This disassembly is mediated by another protein called HSC70 (yellow), and its cofactor protein auxilin (orange).
Janet Iwasa, University of Utah
View Media
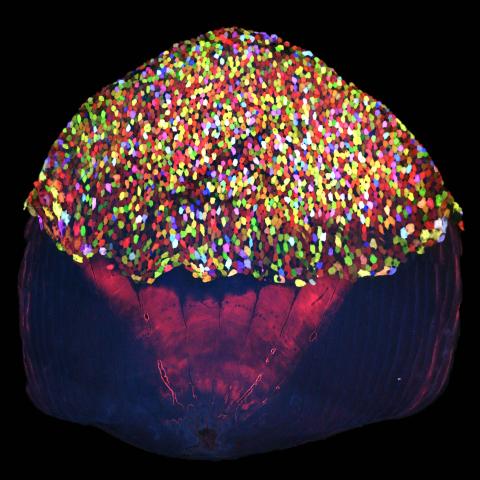
3783: A multicolored fish scale 2
3783: A multicolored fish scale 2
Each of the tiny colored specs in this image is a cell on the surface of a fish scale. To better understand how wounds heal, scientists have inserted genes that make cells brightly glow in different colors into the skin cells of zebrafish, a fish often used in laboratory research. The colors enable the researchers to track each individual cell, for example, as it moves to the location of a cut or scrape over the course of several days. These technicolor fish endowed with glowing skin cells dubbed "skinbow" provide important insight into how tissues recover and regenerate after an injury.
For more information on skinbow fish, see the Biomedical Beat blog post Visualizing Skin Regeneration in Real Time and a press release from Duke University highlighting this research. Related to image 3782.
For more information on skinbow fish, see the Biomedical Beat blog post Visualizing Skin Regeneration in Real Time and a press release from Duke University highlighting this research. Related to image 3782.
Chen-Hui Chen and Kenneth Poss, Duke University
View Media
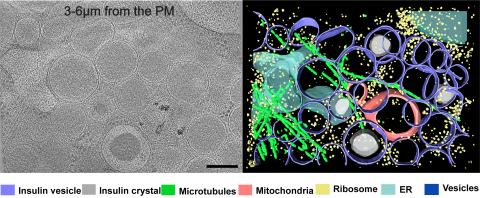
6607: Cryo-ET cell cross-section visualizing insulin vesicles
6607: Cryo-ET cell cross-section visualizing insulin vesicles
On the left, a cross-section slice of a rat pancreas cell captured using cryo-electron tomography (cryo-ET). On the right, a color-coded, 3D version of the image highlighting cell structures. Visible features include insulin vesicles (purple rings), insulin crystals (gray circles), microtubules (green rods), ribosomes (small yellow circles). The black line at the bottom right of the left image represents 200 nm. Related to image 6608.
Xianjun Zhang, University of Southern California.
View Media
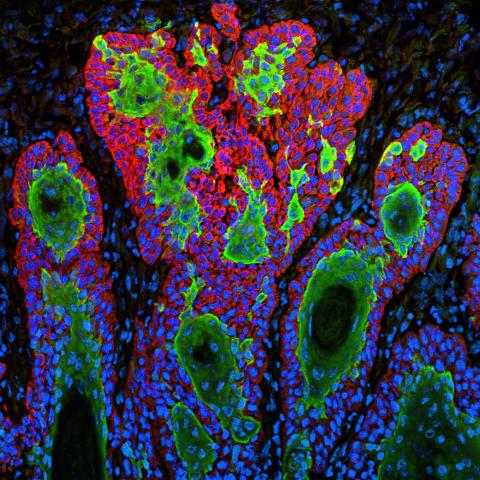
3628: Skin cancer cells (squamous cell carcinoma)
3628: Skin cancer cells (squamous cell carcinoma)
This image shows the uncontrolled growth of cells in squamous cell carcinoma, the second most common form of skin cancer. If caught early, squamous cell carcinoma is usually not life-threatening.
This image was part of the Life: Magnified exhibit that ran from June 3, 2014, to January 21, 2015, at Dulles International Airport.
This image was part of the Life: Magnified exhibit that ran from June 3, 2014, to January 21, 2015, at Dulles International Airport.
Markus Schober and Elaine Fuchs, The Rockefeller University
View Media
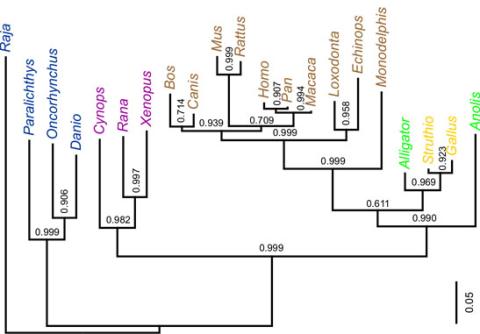
2474: Dinosaur evolutionary tree
2474: Dinosaur evolutionary tree
Analysis of 68 million-year-old collagen molecule fragments preserved in a T. rex femur confirmed what paleontologists have said for decades: Dinosaurs are close relatives of chickens, ostriches, and to a lesser extent, alligators. A Harvard University research team, including NIGMS-supported postdoctoral research fellow Chris Organ, used sophisticated statistical and computational tools to compare the ancient protein to ones from 21 living species. Because evolutionary processes produce similarities across species, the methods and results may help illuminate other areas of the evolutionary tree. Featured in the May 21, 2008 Biomedical Beat.
Chris Organ, Harvard University
View Media
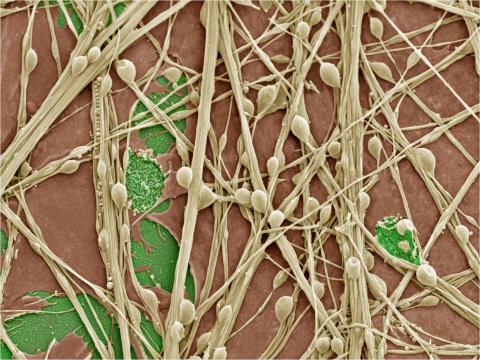
3399: Synapses in culture
3399: Synapses in culture
Cultured hippocampal neurons grown on a substrate of glial cells (astrocytes). The glial cells form the pink/brown underlayment in this image. The tan threads are the neurons. The round tan balls are synapses, the points where neurons meet and communicate with each other. The cover slip underlying the cells is green. Neurons in culture can be used to study synaptic plasticity, activity-dependent protein turnover, and other topics in neuroscience.
National Center for Microscopy and Imaging Research
View Media
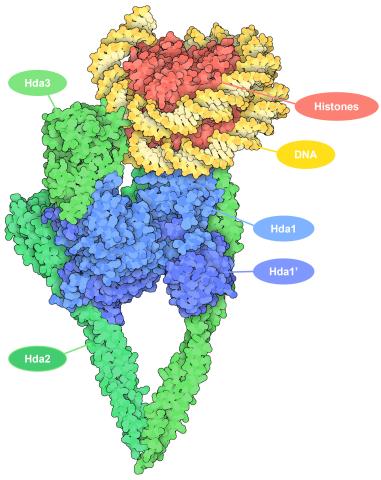
7001: Histone deacetylases
7001: Histone deacetylases
The human genome contains much of the information needed for every cell in the body to function. However, different types of cells often need different types of information. Access to DNA is controlled, in part, by how tightly it’s wrapped around proteins called histones to form nucleosomes. The complex shown here, from yeast cells (PDB entry 6Z6P), includes several histone deacetylase (HDAC) enzymes (green and blue) bound to a nucleosome (histone proteins in red; DNA in yellow). The yeast HDAC enzymes are similar to the human enzymes. Two enzymes form a V-shaped clamp (green) that holds the other others, a dimer of the Hda1 enzymes (blue). In this assembly, Hda1 is activated and positioned to remove acetyl groups from histone tails.
Amy Wu and Christine Zardecki, RCSB Protein Data Bank.
View Media
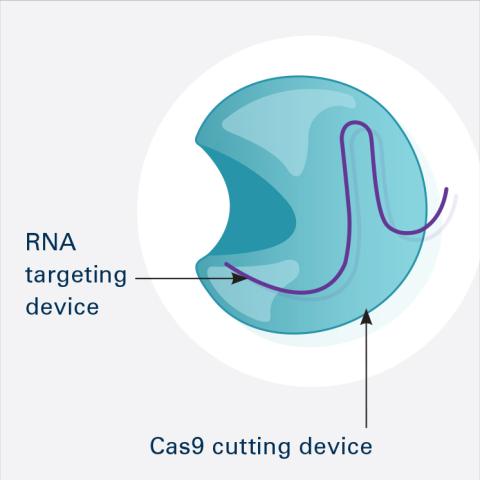
6465: CRISPR Illustration Frame 1
6465: CRISPR Illustration Frame 1
This illustration shows, in simplified terms, how the CRISPR-Cas9 system can be used as a gene-editing tool. This is the first frame in a series of four. The CRISPR system has two components joined together: a finely tuned targeting device (a small strand of RNA programmed to look for a specific DNA sequence) and a strong cutting device (an enzyme called Cas9 that can cut through a double strand of DNA).
For an explanation and overview of the CRISPR-Cas9 system, see the iBiology video, and find the full CRIPSR illustration here.
For an explanation and overview of the CRISPR-Cas9 system, see the iBiology video, and find the full CRIPSR illustration here.
National Institute of General Medical Sciences.
View Media
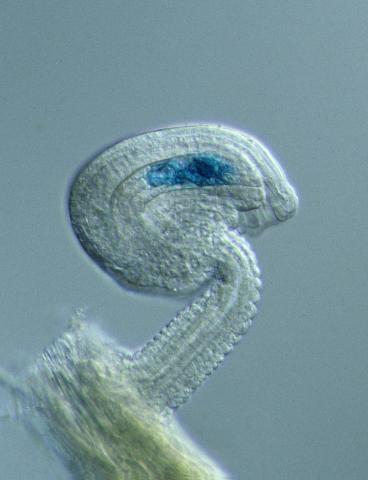
2418: Genetic imprinting in Arabidopsis
2418: Genetic imprinting in Arabidopsis
This delicate, birdlike projection is an immature seed of the Arabidopsis plant. The part in blue shows the cell that gives rise to the endosperm, the tissue that nourishes the embryo. The cell is expressing only the maternal copy of a gene called MEDEA. This phenomenon, in which the activity of a gene can depend on the parent that contributed it, is called genetic imprinting. In Arabidopsis, the maternal copy of MEDEA makes a protein that keeps the paternal copy silent and reduces the size of the endosperm. In flowering plants and mammals, this sort of genetic imprinting is thought to be a way for the mother to protect herself by limiting the resources she gives to any one embryo. Featured in the May 16, 2006, issue of Biomedical Beat.
Robert Fischer, University of California, Berkeley
View Media
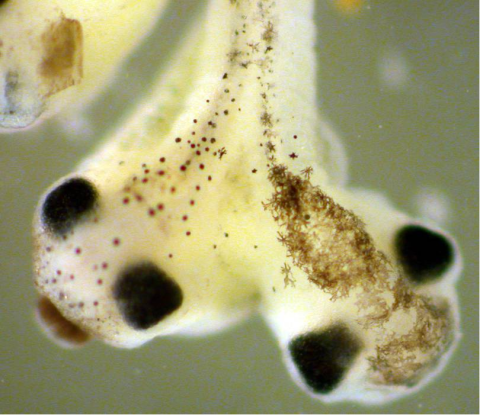
2755: Two-headed Xenopus laevis tadpole
2755: Two-headed Xenopus laevis tadpole
Xenopus laevis, the African clawed frog, has long been used as a research organism for studying embryonic development. The abnormal presence of RNA encoding the signaling molecule plakoglobin causes atypical signaling, giving rise to a two-headed tadpole.
Michael Klymkowsky, University of Colorado, Boulder
View Media
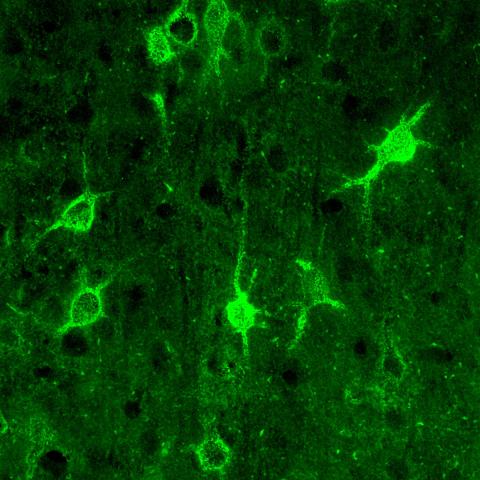
3741: Confocal microscopy of perineuronal nets in the brain 1
3741: Confocal microscopy of perineuronal nets in the brain 1
The photo shows a confocal microscopy image of perineuronal nets (PNNs), which are specialized extracellular matrix (ECM) structures in the brain. The PNN surrounds some nerve cells in brain regions including the cortex, hippocampus and thalamus. Researchers study the PNN to investigate their involvement stabilizing the extracellular environment and forming nets around nerve cells and synapses in the brain. Abnormalities in the PNNs have been linked to a variety of disorders, including epilepsy and schizophrenia, and they limit a process called neural plasticity in which new nerve connections are formed. To visualize the PNNs, researchers labeled them with Wisteria floribunda agglutinin (WFA)-fluorescein. Related to image 3742.
Tom Deerinck, National Center for Microscopy and Imaging Research (NCMIR)
View Media
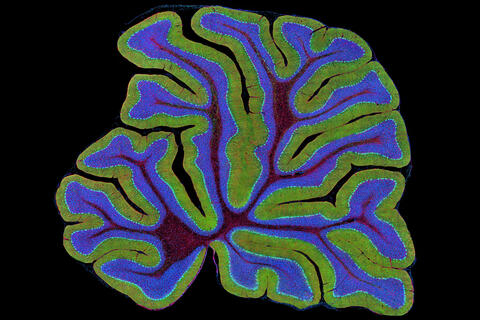
3723: Fluorescent microscopy of kidney tissue
3723: Fluorescent microscopy of kidney tissue
Serum albumin (SA) is the most abundant protein in the blood plasma of mammals. SA has a characteristic heart-shape structure and is a highly versatile protein. It helps maintain normal water levels in our tissues and carries almost half of all calcium ions in human blood. SA also transports some hormones, nutrients and metals throughout the bloodstream. Despite being very similar to our own SA, those from other animals can cause some mild allergies in people. Therefore, some scientists study SAs from humans and other mammals to learn more about what subtle structural or other differences cause immune responses in the body.
Related to entries 3725 and 3675.
Related to entries 3725 and 3675.
Tom Deerinck , National Center for Microscopy and Imaging Research
View Media
1083: Natcher Building 03
1083: Natcher Building 03
NIGMS staff are located in the Natcher Building on the NIH campus.
Alisa Machalek, National Institute of General Medical Sciences
View Media

3477: HIV Capsid
3477: HIV Capsid
This image is a computer-generated model of the approximately 4.2 million atoms of the HIV capsid, the shell that contains the virus' genetic material. Scientists determined the exact structure of the capsid and the proteins that it's made of using a variety of imaging techniques and analyses. They then entered these data into a supercomputer that produced the atomic-level image of the capsid. This structural information could be used for developing drugs that target the capsid, possibly leading to more effective therapies. Related to image 6601.
Juan R. Perilla and the Theoretical and Computational Biophysics Group, University of Illinois at Urbana-Champaign
View Media
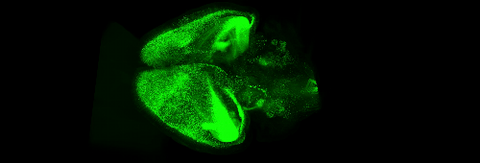
6931: Mouse brain 3
6931: Mouse brain 3
Various views of a mouse brain that was genetically modified so that subpopulations of its neurons glow. Researchers often study mice because they share many genes with people and can shed light on biological processes, development, and diseases in humans.
This video was captured using a light sheet microscope.
Related to images 6929 and 6930.
This video was captured using a light sheet microscope.
Related to images 6929 and 6930.
Prayag Murawala, MDI Biological Laboratory and Hannover Medical School.
View Media
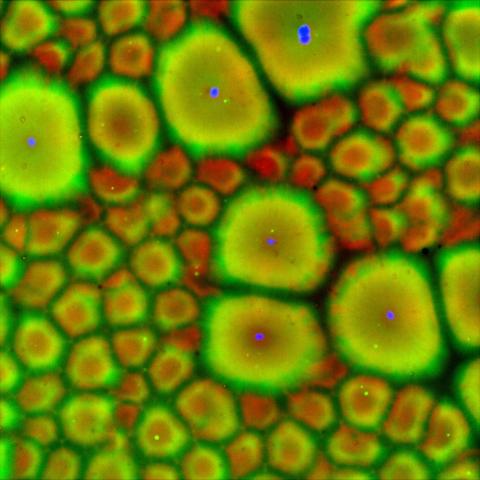
6585: Cell-like compartments from frog eggs 2
6585: Cell-like compartments from frog eggs 2
Cell-like compartments that spontaneously emerged from scrambled frog eggs, with nuclei (blue) from frog sperm. Endoplasmic reticulum (red) and microtubules (green) are also visible. Regions without nuclei formed smaller compartments. Image created using epifluorescence microscopy.
For more photos of cell-like compartments from frog eggs view: 6584, 6586, 6591, 6592, and 6593.
For videos of cell-like compartments from frog eggs view: 6587, 6588, 6589, and 6590.
Xianrui Cheng, Stanford University School of Medicine.
View Media
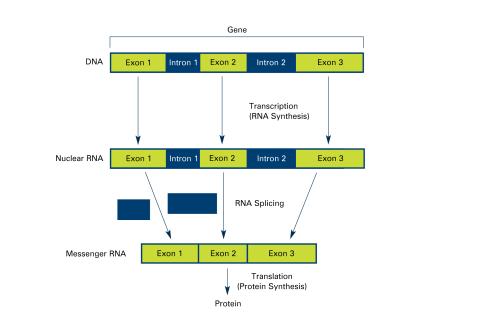
2551: Introns (with labels)
2551: Introns (with labels)
Genes are often interrupted by stretches of DNA (introns, blue) that do not contain instructions for making a protein. The DNA segments that do contain protein-making instructions are known as exons (green). See image 2550 for an unlabeled version of this illustration. Featured in The New Genetics.
Crabtree + Company
View Media
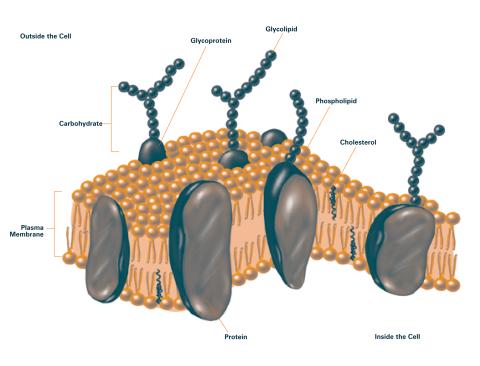
2524: Plasma membrane (with labels)
2524: Plasma membrane (with labels)
The plasma membrane is a cell's protective barrier. See image 2523 for an unlabeled version of this illustration. Featured in The Chemistry of Health.
Crabtree + Company
View Media
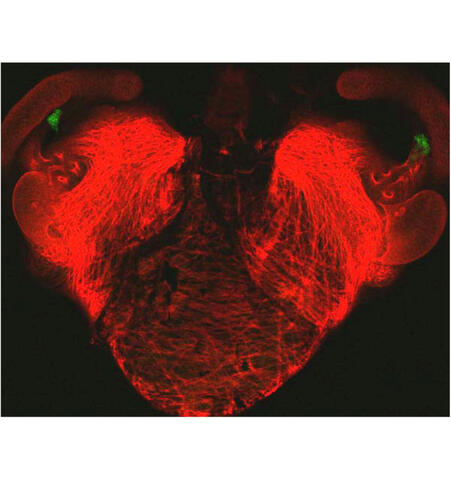
7018: Bacterial cells aggregating above the light organ of the Hawaiian bobtail squid
7018: Bacterial cells aggregating above the light organ of the Hawaiian bobtail squid
A light organ (~0.5 mm across) of a juvenile Hawaiian bobtail squid, Euprymna scolopes. Movement of cilia on the surface of the organ aggregates bacterial symbionts (green) into two areas above sets of pores that lead to interior crypts. This image was taken using a confocal fluorescence microscope.
Related to images 7016, 7017, 7019, and 7020.
Related to images 7016, 7017, 7019, and 7020.
Margaret J. McFall-Ngai, Carnegie Institution for Science/California Institute of Technology, and Edward G. Ruby, California Institute of Technology.
View Media
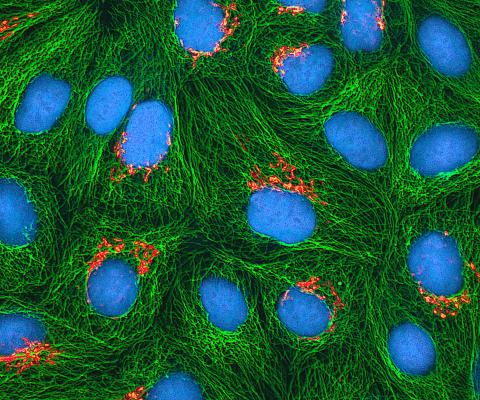
3522: HeLa cells
3522: HeLa cells
Multiphoton fluorescence image of cultured HeLa cells with a fluorescent protein targeted to the Golgi apparatus (orange), microtubules (green) and counterstained for DNA (cyan). Nikon RTS2000MP custom laser scanning microscope. See related images 3518, 3519, 3520, 3521.
National Center for Microscopy and Imaging Research (NCMIR)
View Media
6520: HeLa cell undergoing division into two daughter cells
6520: HeLa cell undergoing division into two daughter cells
Here, a human HeLa cell (a type of immortal cell line used in laboratory experiments) is undergoing cell division. They come from cervical cancer cells that were obtained in 1951 from Henrietta Lacks, a patient at the Johns Hopkins Hospital. The final stage of division, called cytokinesis, occurs after the genomes—shown in yellow—have split into two new daughter cells. The myosin II is a motor protein shown in blue, and the actin filaments, which are types of protein that support cell structure, are shown in red.
Dylan T. Burnette, Ph.D., Vanderbilt University School of Medicine.
View Media
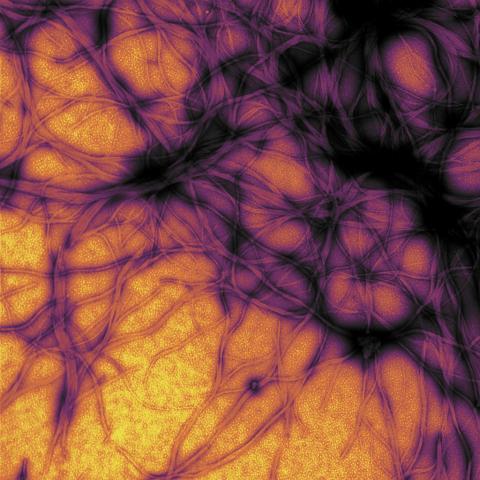
3460: Prion protein fibrils 1
3460: Prion protein fibrils 1
Recombinant proteins such as the prion protein shown here are often used to model how proteins misfold and sometimes polymerize in neurodegenerative disorders. This prion protein was expressed in E. coli, purified and fibrillized at pH 7. Image taken in 2004 for a research project by Roger Moore, Ph.D., at Rocky Mountain Laboratories that was published in 2007 in Biochemistry. This image was not used in the publication.
Ken Pekoc (public affairs officer) and Julie Marquardt, NIAID/ Rocky Mountain Laboratories
View Media
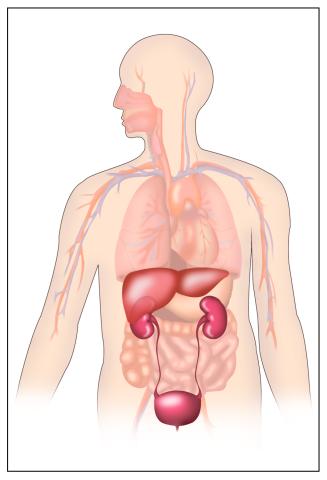
2496: Body toxins
2496: Body toxins
Body organs such as the liver and kidneys process chemicals and toxins. These "target" organs are susceptible to damage caused by these substances. See image 2497 for a labeled version of this illustration.
Crabtree + Company
View Media
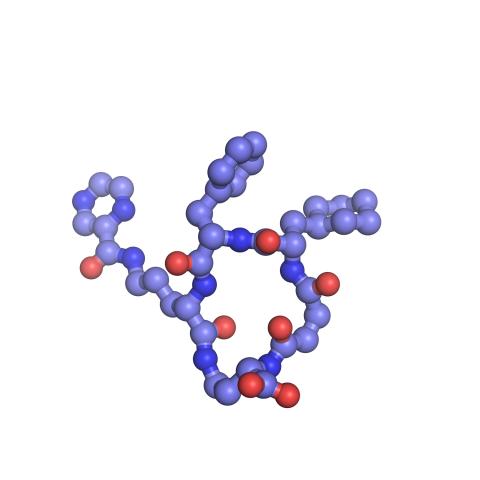
3418: X-ray co-crystal structure of Src kinase bound to a DNA-templated macrocycle inhibitor 6
3418: X-ray co-crystal structure of Src kinase bound to a DNA-templated macrocycle inhibitor 6
X-ray co-crystal structure of Src kinase bound to a DNA-templated macrocycle inhibitor. Related to images 3413, 3414, 3415, 3416, 3417, and 3419.
Markus A. Seeliger, Stony Brook University Medical School and David R. Liu, Harvard University
View Media
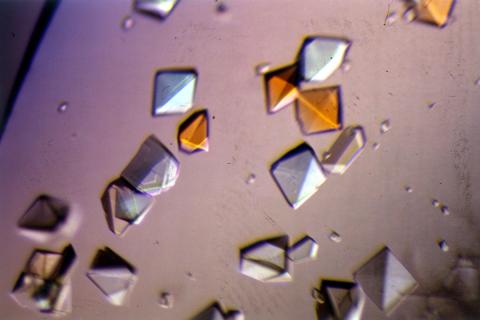
2412: Pig alpha amylase
2412: Pig alpha amylase
Crystals of porcine alpha amylase protein created for X-ray crystallography, which can reveal detailed, three-dimensional protein structures.
Alex McPherson, University of California, Irvine
View Media

1060: Protein crystals
1060: Protein crystals
Structural biologists create crystals of proteins, shown here, as a first step in a process called X-ray crystallography, which can reveal detailed, three-dimensional protein structures.
Alex McPherson, University of California, Irvine
View Media
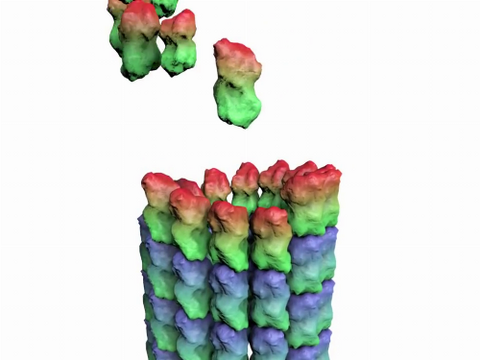
3650: How a microtubule builds and deconstructs
3650: How a microtubule builds and deconstructs
A microtubule, part of the cell's skeleton, builds and deconstructs.
View Media
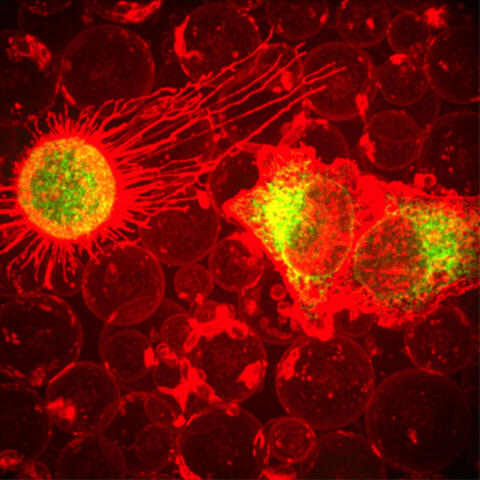
5887: Plasma-Derived Membrane Vesicles
5887: Plasma-Derived Membrane Vesicles
This fiery image doesn’t come from inside a bubbling volcano. Instead, it shows animal cells caught in the act of making bubbles, or blebbing. Some cells regularly pinch off parts of their membranes to produce bubbles filled with a mix of proteins and fats. The bubbles (red) are called plasma-derived membrane vesicles, or PMVs, and can travel to other parts of the body where they may aid in cell-cell communication. The University of Texas, Austin, researchers responsible for this photo are exploring ways to use PMVs to deliver medicines to precise locations in the body.
This image, entered in the Biophysical Society’s 2017 Art of Science Image contest, used two-channel spinning disk confocal fluorescence microscopy. It was also featured in the NIH Director’s Blog in May 2017.
This image, entered in the Biophysical Society’s 2017 Art of Science Image contest, used two-channel spinning disk confocal fluorescence microscopy. It was also featured in the NIH Director’s Blog in May 2017.
Jeanne Stachowiak, University of Texas at Austin
View Media
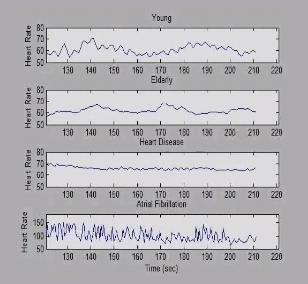
3596: Heart rates time series image
3596: Heart rates time series image
These time series show the heart rates of four different individuals. Automakers use steel scraps to build cars, construction companies repurpose tires to lay running tracks, and now scientists are reusing previously discarded medical data to better understand our complex physiology. Through a website called PhysioNet developed in part by Beth Israel Deaconess Medical Center cardiologist Ary Goldberger, scientists can access complete physiologic recordings, such as heart rate, respiration, brain activity and gait. They then can use free software to analyze the data and find patterns in it. The patterns could ultimately help health care professionals diagnose and treat health conditions like congestive heart failure, sleeping disorders, epilepsy and walking problems. PhysioNet is supported by NIH's National Institute of Biomedical Imaging and Bioengineering as well as by NIGMS.
Madalena Costa and Ary Goldberger, Beth Israel Deaconess Medical Center
View Media
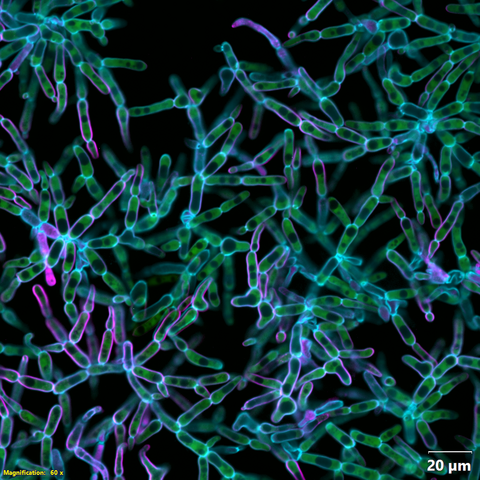
6970: Snowflake yeast 2
6970: Snowflake yeast 2
Multicellular yeast called snowflake yeast that researchers created through many generations of directed evolution from unicellular yeast. Cells are connected to one another by their cell walls, shown in blue. Stained cytoplasm (green) and membranes (magenta) show that the individual cells remain separate. This image was captured using spinning disk confocal microscopy.
Related to images 6969 and 6971.
Related to images 6969 and 6971.
William Ratcliff, Georgia Institute of Technology.
View Media
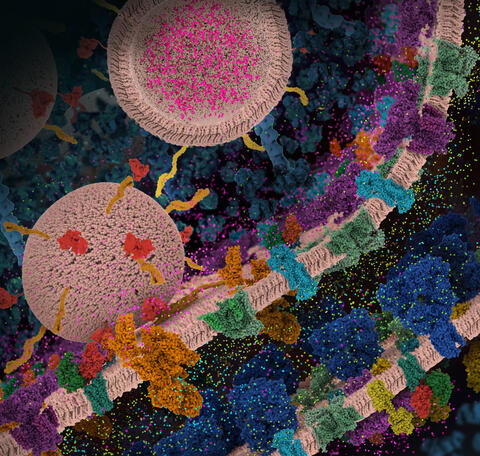
6992: Molecular view of glutamatergic synapse
6992: Molecular view of glutamatergic synapse
This illustration highlights spherical pre-synaptic vesicles that carry the neurotransmitter glutamate. The presynaptic and postsynaptic membranes are shown with proteins relevant for transmitting and modulating the neuronal signal.
PDB 101’s Opioids and Pain Signaling video explains how glutamatergic synapses are involved in the process of pain signaling.
PDB 101’s Opioids and Pain Signaling video explains how glutamatergic synapses are involved in the process of pain signaling.
Amy Wu and Christine Zardecki, RCSB Protein Data Bank.
View Media
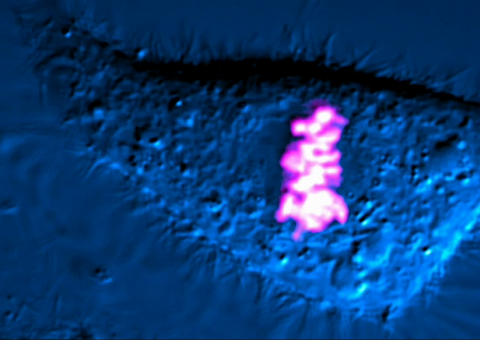
6965: Dividing cell
6965: Dividing cell
As this cell was undergoing cell division, it was imaged with two microscopy techniques: differential interference contrast (DIC) and confocal. The DIC view appears in blue and shows the entire cell. The confocal view appears in pink and shows the chromosomes.
Dylan T. Burnette, Vanderbilt University School of Medicine.
View Media

2360: Cell-free protein synthesizers
2360: Cell-free protein synthesizers
Both instruments shown were developed by CellFree Sciences of Yokohama, Japan. The instrument on the left, the GeneDecoder 1000, can generate 384 proteins from their corresponding genes, or gene fragments, overnight. It is used to screen for properties such as level of protein production and degree of solubility. The instrument on the right, the Protemist Protein Synthesizer, is used to generate the larger amounts of protein needed for protein structure determinations.
Center for Eukaryotic Structural Genomics
View Media
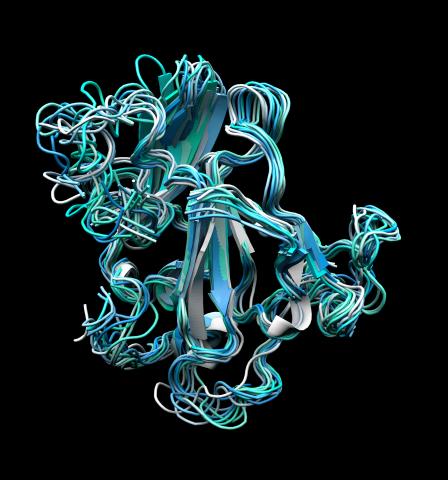
5866: Structure of a key antigen protein involved with Hepatitis C Virus infection
5866: Structure of a key antigen protein involved with Hepatitis C Virus infection
A three-dimensional representation of the structure of E2, a key antigen protein involved with hepatitis C virus infection.
Mansun Law Associate Professor Department of Immunolgy and Microbial Science The Scripps Research Institute
View Media

2579: Bottles of warfarin
2579: Bottles of warfarin
In 2007, the FDA modified warfarin's label to indicate that genetic makeup may affect patient response to the drug. The widely used blood thinner is sold under the brand name Coumadin®. Scientists involved in the NIH Pharmacogenetics Research Network are investigating whether genetic information can be used to improve optimal dosage prediction for patients.
Alisa Machalek, NIGMS/NIH
View Media
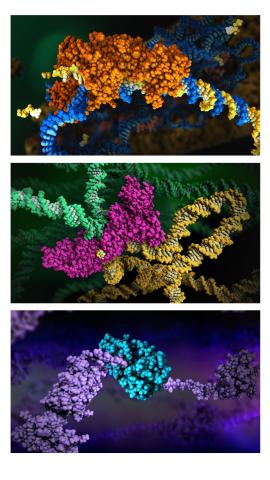
6999: HIV enzyme
6999: HIV enzyme
These images model the molecular structures of three enzymes with critical roles in the life cycle of the human immunodeficiency virus (HIV). At the top, reverse transcriptase (orange) creates a DNA copy (yellow) of the virus's RNA genome (blue). In the middle image, integrase (magenta) inserts this DNA copy in the DNA genome (green) of the infected cell. At the bottom, much later in the viral life cycle, protease (turquoise) chops up a chain of HIV structural protein (purple) to generate the building blocks for making new viruses. See these enzymes in action on PDB 101’s video A Molecular View of HIV Therapy.
Amy Wu and Christine Zardecki, RCSB Protein Data Bank.
View Media

3526: 800 MHz NMR magnet
3526: 800 MHz NMR magnet
Scientists use nuclear magnetic spectroscopy (NMR) to determine the detailed, 3D structures of molecules.
Asokan Anbanandam, University of Kansas
View Media

2371: NMR spectrometer
2371: NMR spectrometer
This photo shows a Varian Unity Inova 900 MHz, 21.1 T standard bore magnet Nuclear Magnetic Resonnance (NMR) spectrometer. NMR spectroscopy provides data used to determine the structures of proteins in solution, rather than in crystal form, as in X-ray crystallography. The technique is limited to smaller proteins or protein fragments in a high throughput approach.
Center for Eukaryotic Structural Genomics
View Media
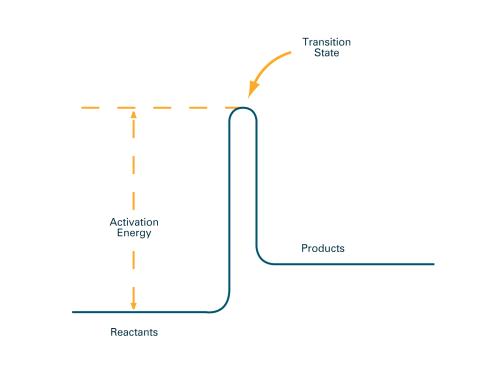
2526: Activation energy (with labels)
2526: Activation energy (with labels)
To become products, reactants must overcome an energy hill. See image 2525 for an unlabeled version of this illustration. Featured in The Chemistry of Health.
Crabtree + Company
View Media
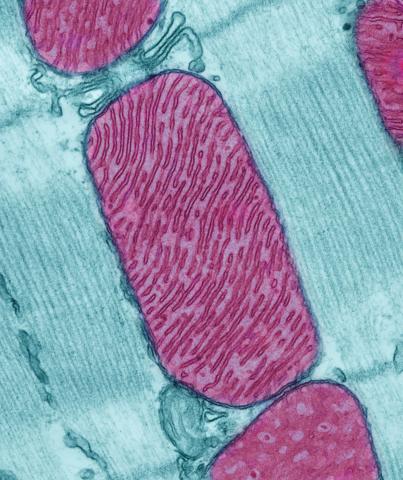
3661: Mitochondria from rat heart muscle cell
3661: Mitochondria from rat heart muscle cell
These mitochondria (red) are from the heart muscle cell of a rat. Mitochondria have an inner membrane that folds in many places (and that appears here as striations). This folding vastly increases the surface area for energy production. Nearly all our cells have mitochondria. Related to image 3664.
National Center for Microscopy and Imaging Research
View Media
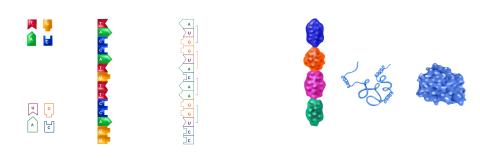
2509: From DNA to Protein
2509: From DNA to Protein
Nucleotides in DNA are copied into RNA, where they are read three at a time to encode the amino acids in a protein. Many parts of a protein fold as the amino acids are strung together.
See image 2510 for a labeled version of this illustration.
Featured in The Structures of Life.
See image 2510 for a labeled version of this illustration.
Featured in The Structures of Life.
Crabtree + Company
View Media
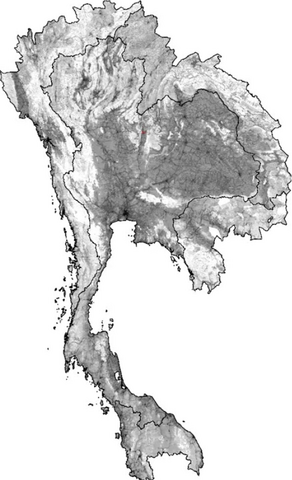
2574: Simulation of uncontrolled avian flu outbreak
2574: Simulation of uncontrolled avian flu outbreak
This video simulation shows what an uncontrolled outbreak of transmissible avian flu among people living in Thailand might look like. Red indicates new cases while green indicates areas where the epidemic has finished. The video shows the spread of infection and recovery over 300 days in Thailand and neighboring countries.
Neil M. Ferguson, Imperial College London
View Media
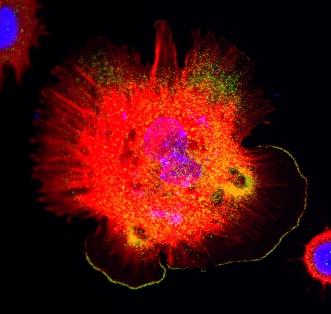
3328: Spreading Cells 01
3328: Spreading Cells 01
Cells move forward with lamellipodia and filopodia supported by networks and bundles of actin filaments. Proper, controlled cell movement is a complex process. Recent research has shown that an actin-polymerizing factor called the Arp2/3 complex is the key component of the actin polymerization engine that drives amoeboid cell motility. ARPC3, a component of the Arp2/3 complex, plays a critical role in actin nucleation. In this photo, the ARPC3+/+ fibroblast cells were fixed and stained with Alexa 546 phalloidin for F-actin (red), Arp2 (green), and DAPI to visualize the nucleus (blue). Arp2, a subunit of the Arp2/3 complex, is localized at the lamellipodia leading edge of ARPC3+/+ fibroblast cells. Related to images 3329, 3330, 3331, 3332, and 3333.
Rong Li and Praveen Suraneni, Stowers Institute for Medical Research
View Media
