Switch to Gallery View
Image and Video Gallery
This is a searchable collection of scientific photos, illustrations, and videos. The images and videos in this gallery are licensed under Creative Commons Attribution Non-Commercial ShareAlike 3.0. This license lets you remix, tweak, and build upon this work non-commercially, as long as you credit and license your new creations under identical terms.
Weblike sheath covering developing egg chambers in a giant grasshopper
3616
The lubber grasshopper, found throughout the southern United States, is frequently used in biology classes to teach students about the respiratory system of insects. Kevin Edwards, Johny Shajahan, and Doug Whitman, Illinois State University. View MediaKinesin moves cellular cargo
3491
A protein called kinesin (blue) is in charge of moving cargo around inside cells and helping them divide. Charles Sindelar, Yale University View MediaProtein from Methanobacterium thermoautotrophicam
2374
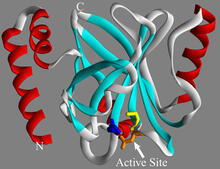
A knotted protein from an archaebacterium called Methanobacterium thermoautotrophicam. This organism breaks down waste products and produces methane gas. Midwest Center For Structural Genomics, PSI View MediaEnzymes convert subtrates into products (with labels)
2522
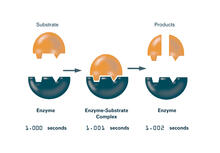
Enzymes convert substrates into products very quickly. See image 2521 for an unlabeled version of this illustration. Featured in The Chemistry of Health. Crabtree + Company View MediaTrajectories of labeled cell receptors
2801
Trajectories of single molecule labeled cell surface receptors. This is an example of NIH-supported research on single-cell analysis. Gaudenz Danuser, Harvard Medical School View MediaSeeing signaling protein activation in cells 04
2454
Cdc42, a member of the Rho family of small guanosine triphosphatase (GTPase) proteins, regulates multiple cell functions, including motility, proliferation, apoptosis, and cell morphology. Klaus Hahn, University of North Carolina, Chapel Hill Medical School View MediaPig trypsin crystal
2403
A crystal of pig trypsin protein created for X-ray crystallography, which can reveal detailed, three-dimensional protein structures. Alex McPherson, University of California, Irvine View MediaMagnesium transporter protein from E. faecalis
2345
Structure of a magnesium transporter protein from an antibiotic-resistant bacterium (Enterococcus faecalis) found in the human gut. New York Structural GenomiX Consortium View MediaPig trypsin (2)
2413
A crystal of porcine trypsin protein created for X-ray crystallography, which can reveal detailed, three-dimensional protein structures. Alex McPherson, University of California, Irvine View MediaDisrupted vascular development in frog embryos
3403
Disassembly of vasculature in kdr:GFP frogs following addition of 250 µM TBZ. Related to images 3404 and 3505. Hye Ji Cha, University of Texas at Austin View MediaRibbon diagram of a cefotaxime-CCD-1 complex
6766
CCD-1 is an enzyme produced by the bacterium Clostridioides difficile that helps it resist antibiotics. Keith Hodgson, Stanford University. View MediaRNA polymerase
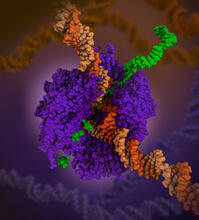
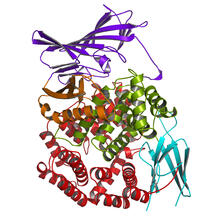
6993
RNA polymerase (purple) is a complex enzyme at the heart of transcription. Amy Wu and Christine Zardecki, RCSB Protein Data Bank. View MediaMouse brain slice showing nerve cells
6901
A 20-µm thick section of mouse midbrain. The nerve cells are transparent and weren’t stained. Michael Shribak, Marine Biological Laboratory/University of Chicago. View MediaAtomic-level structure of the HIV capsid
6601
This animation shows atoms of the HIV capsid, the shell that encloses the virus's genetic material. Juan R. Perilla and the Theoretical and Computational Biophysics Group, University of Illinois at Urbana-Champaign View MediaG switch
2536
The G switch allows our bodies to respond rapidly to hormones. See images 2537 and 2538 for labeled versions of this image. Crabtree + Company View MediaHimastatin, 360-degree view
6851
A 360-degree view of the molecule himastatin, which was first isolated from the bacterium Streptomyces himastatinicus. Himastatin shows antibiotic activity. Mohammad Movassaghi, Massachusetts Institute of Technology. View MediaSphingolipid S1P1 receptor
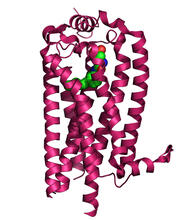
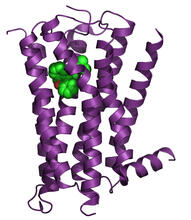
3362
The receptor is shown bound to an antagonist, ML056. Raymond Stevens, The Scripps Research Institute View MediaH1 histamine receptor
3360
The receptor is shown bound to an inverse agonist, doxepin. Raymond Stevens, The Scripps Research Institute View MediaEarly ribbon drawing of a protein
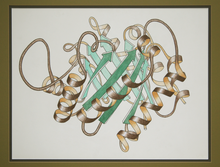
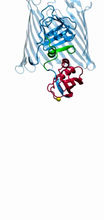
2748
This ribbon drawing of a protein hand drawn and colored by researcher Jane Richardson in 1981 helped originate the ribbon representation of proteins that is now ubiquitous in molecular graphics. Jane Richardson, Duke University Medical Center View MediaTransient receptor potential channel TRPV5
6577
A 3D reconstruction of a transient receptor potential channel called TRPV5 that was created based on cryo-electron microscopy images. Vera Moiseenkova-Bell, University of Pennsylvania. View MediaDrosophila (fruit fly) myosin 1D motility assay
6562
Actin gliding powered by myosin 1D. Note the counterclockwise motion of the gliding actin filaments. Serapion Pyrpassopoulos and E. Michael Ostap, University of Pennsylvania View MediaMeasles virus
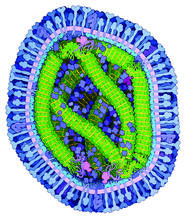
6995
A cross section of the measles virus in which six proteins work together to infect cells. The measles virus is extremely infectious; 9 out of 10 people exposed will contract the disease. Amy Wu and Christine Zardecki, RCSB Protein Data Bank. View MediaAminopeptidase N from N. meningitidis
2341
Model of the enzyme aminopeptidase N from the human pathogen Neisseria meningitidis, which can cause meningitis epidemics. Midwest Center for Structural Genomics, PSI View MediaCytonemes in developing fruit fly cells
3574
Scientists have long known that multicellular organisms use biological molecules produced by one cell and sensed by another to transmit messages that, for instance, guide proper development of organs Sougata Roy, University of California, San Francisco View MediaBacteria working to eat
2304
Gram-negative bacteria perform molecular acrobatics just to eat. Because they're encased by two membranes, they must haul nutrients across both. Emad Tajkhorshid, University of Illinois at Urbana-Champaign View MediaStetten Lecture 2017poster image
5896
This image is featured on the poster for Dr. Rommie Amaro's 2017 Stetten Lecture. Dr. Rommie Amaro, University of California, San Diego View Media3D reconstruction of a tubular matrix in peripheral endoplasmic reticulum
5857
Detailed three-dimensional reconstruction of a tubular matrix in a thin section of the peripheral endoplasmic reticulum between the plasma membranes of the cell. Jennifer Lippincott-Schwartz, Howard Hughes Medical Institute Janelia Research Campus, Virginia View MediaRNA Polymerase II
2484
NIGMS-funded researchers led by Roger Kornberg solved the structure of RNA polymerase II. David Bushnell, Ken Westover and Roger Kornberg, Stanford University View MediaHistones in chromatin (with labels)
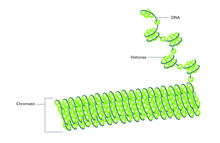
2561
Histone proteins loop together with double-stranded DNA to form a structure that resembles beads on a string. Crabtree + Company View MediaTFIID complex binds DNA to start gene transcription
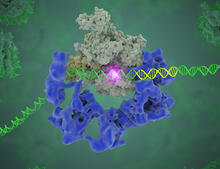
3766
Gene transcription is a process by which the genetic information encoded in DNA is transcribed into RNA. Eva Nogales, Berkeley Lab View MediaDiversity oriented synthesis: generating skeletal diversity using folding processes
3327
This 1 1/2-minute video animation was produced for chemical biologist Stuart Schreiber's lab page. The animation shows how diverse chemical structures can be produced in the lab. Eric Keller View MediaIsolated Planarian Pharynx
3593
The feeding tube, or pharynx, of a planarian worm with cilia shown in red and muscle fibers shown in green View MediaSortase b from B. anthracis
2386
Structure of sortase b from the bacterium B. anthracis, which causes anthrax. Sortase b is an enzyme used to rob red blood cells of iron, which the bacteria need to survive. Midwest Center for Structural Genomics, PSI View MediaAssembly of the HIV capsid
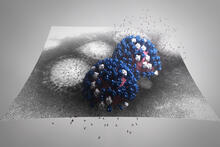
5729
The HIV capsid is a pear-shaped structure that is made of proteins the virus needs to mature and become infective. John Grime and Gregory Voth, The University of Chicago View MediaJack bean concanavalin A
2407
Crystals of jack bean concanavalin A protein created for X-ray crystallography, which can reveal detailed, three-dimensional protein structures. Alex McPherson, University of California, Irvine View MediaTiny strands of tubulin, a protein in a cell's skeleton
3611
Just as our bodies rely on bones for structural support, our cells rely on a cellular skeleton. Pakorn Kanchanawong, National University of Singapore and National Heart, Lung, and Blood Institute, National Institutes of Health; and Clare Waterman, National Heart, Lung, and Blood Institute, National Institutes of Health View MediaHuman Adenovirus
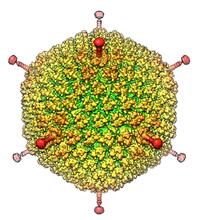
6347
The cryo-EM structure of human adenovirus D26 (HAdV-D26) at near atomic resolution (3.7 Å), determined in collaboration with the NRAMM facility*. National Resource for Automated Molecular Microscopy http://nramm.nysbc.org/nramm-images/ Source: Bridget Carragher View MediaMolecular view of glutamatergic synapse
6992
This illustration highlights spherical pre-synaptic vesicles that carry the neurotransmitter glutamate. Amy Wu and Christine Zardecki, RCSB Protein Data Bank. View MediaAntitoxin GhoS (Illustration 2)
3428
Structure of the bacterial antitoxin protein GhoS. GhoS inhibits the production of a bacterial toxin, GhoT, which can contribute to antibiotic resistance. Rebecca Page and Wolfgang Peti, Brown University and Thomas K. Wood, Pennsylvania State University View MediaMost abundant protein in M. tuberculosis
2378
Model of a protein, antigen 85B, that is the most abundant protein exported by Mycobacterium tuberculosis, which causes most cases of tuberculosis. Mycobacterium Tuberculosis Center, PSI View MediaBacterial alpha amylase
2401
A crystal of bacterial alpha amylase protein created for X-ray crystallography, which can reveal detailed, three-dimensional protein structures. Alex McPherson, University of California, Irvine View MediaCas4 nuclease protein structure
3720
This wreath represents the molecular structure of a protein, Cas4, which is part of a system, known as CRISPR, that bacteria use to protect themselves against viral invaders. Fred Dyda, NIDDK View MediaHeat shock protein complex from Methanococcus jannaschii
2385
Model based on X-ray crystallography of the structure of a small heat shock protein complex from the bacteria, Methanococcus jannaschii. Berkeley Structural Genomics Center, PSI-1 View MediaHen egg lysozyme (2)
2406
A crystal of hen egg lysozyme protein created for X-ray crystallography, which can reveal detailed, three-dimensional protein structures. Alex McPherson, University of California, Irvine View MediaKinases
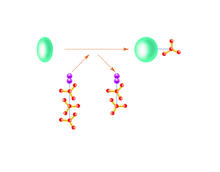
2534
Kinases are enzymes that add phosphate groups (red-yellow structures) to proteins (green), assigning the proteins a code. Crabtree + Company View MediaX-ray co-crystal structure of Src kinase bound to a DNA-templated macrocycle inhibitor 4
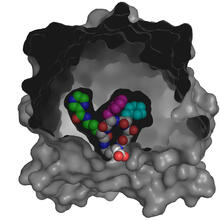
3416
X-ray co-crystal structure of Src kinase bound to a DNA-templated macrocycle inhibitor. Markus A. Seeliger, Stony Brook University Medical School and David R. Liu, Harvard University View MediaTex protein
2338
Model of a member from the Tex protein family, which is implicated in transcriptional regulation and highly conserved in eukaryotes and prokaryotes. New York Structural GenomiX Research Consortium, PSI View MediaHsp33 figure 1
3354
Featured in the March 15, 2012 issue of Biomedical Beat. Related to Hsp33 Figure 2, image 3355. Ursula Jakob and Dana Reichmann, University of Michigan View MediaCalcium uptake during ATP production in mitochondria
3449
Living primary mouse embryonic fibroblasts. Mitochondria (green) stained with the mitochondrial membrane potential indicator, rhodamine 123. Nuclei (blue) are stained with DAPI. Lili Guo, Perelman School of Medicine, University of Pennsylvania View MediaDNA replication origin recognition complex (ORC)
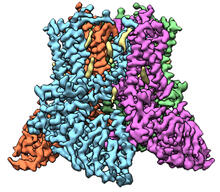
3597
A study published in March 2012 used cryo-electron microscopy to determine the structure of the DNA replication origin recognition complex (ORC), a semi-circular, protein complex (yellow) that recogni Huilin Li, Brookhaven National Laboratory View Media