Switch to Gallery View
Image and Video Gallery
This is a searchable collection of scientific photos, illustrations, and videos. The images and videos in this gallery are licensed under Creative Commons Attribution Non-Commercial ShareAlike 3.0. This license lets you remix, tweak, and build upon this work non-commercially, as long as you credit and license your new creations under identical terms.
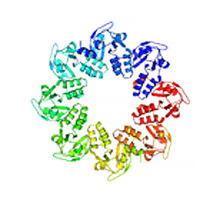
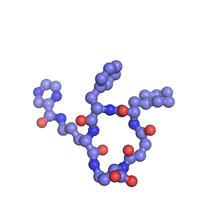
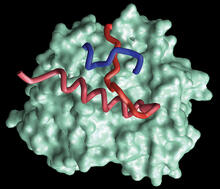
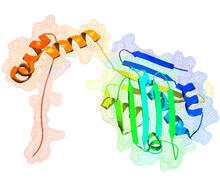
Self-organizing proteins
2771
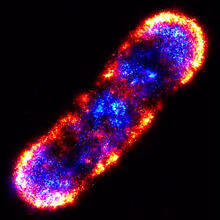
Under the microscope, an E. coli cell lights up like a fireball. Each bright dot marks a surface protein that tells the bacteria to move toward or away from nearby food and toxins. View MediaMouse brain slice showing nerve cells
6901
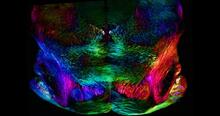
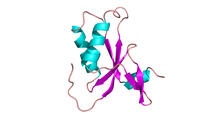
A 20-µm thick section of mouse midbrain. The nerve cells are transparent and weren’t stained. Michael Shribak, Marine Biological Laboratory/University of Chicago. View MediaProtein from Arabidopsis thaliana
2339
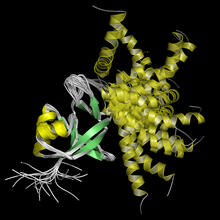
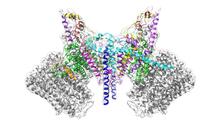
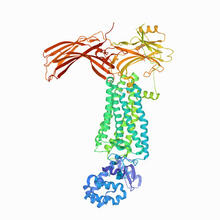
NMR solution structure of a plant protein that may function in host defense. This protein was expressed in a convenient and efficient wheat germ cell-free system. Center for Eukaryotic Structural Genomics View MediaKappa opioid receptor
3359
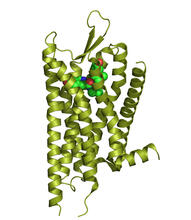
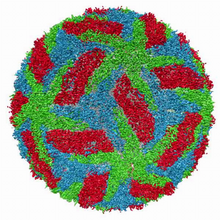
The receptor is shown bound to an antagonist, JDTic. Raymond Stevens, The Scripps Research Institute View MediaJack bean concanavalin A
2407
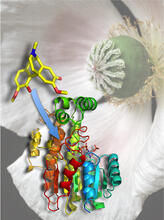
Crystals of jack bean concanavalin A protein created for X-ray crystallography, which can reveal detailed, three-dimensional protein structures. Alex McPherson, University of California, Irvine View MediaAtomic Structure of Poppy Enzyme
3422
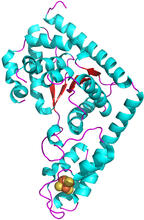
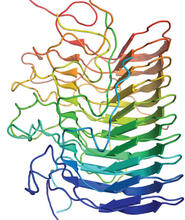
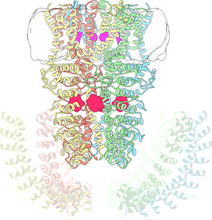
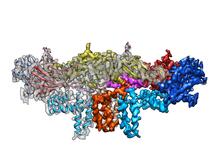
The atomic structure of the morphine biosynthetic enzyme salutaridine reductase bound to the cofactor NADPH. The substrate salutaridine is shown entering the active site. Judy Coyle, Donald Danforth Plant Science Center View MediaTrp_RS - tryptophanyl tRNA-synthetase family of enzymes
2483
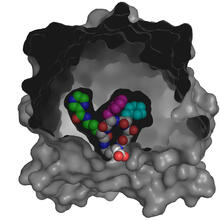
This image represents the structure of TrpRS, a novel member of the tryptophanyl tRNA-synthetase family of enzymes. View MediaX-ray co-crystal structure of Src kinase bound to a DNA-templated macrocycle inhibitor 4
3416
X-ray co-crystal structure of Src kinase bound to a DNA-templated macrocycle inhibitor. Markus A. Seeliger, Stony Brook University Medical School and David R. Liu, Harvard University View MediaPig trypsin (3)
2414
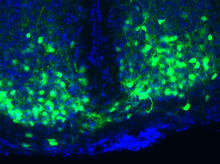
Crystals of porcine trypsin protein created for X-ray crystallography, which can reveal detailed, three-dimensional protein structures. Alex McPherson, University of California, Irvine View MediaMaster clock of the mouse brain
3547
An image of the area of the mouse brain that serves as the 'master clock,' which houses the brain's time-keeping neurons. The nuclei of the clock cells are shown in blue. Erik Herzog, Washington University in St. Louis View MediaHuman opioid receptor structure superimposed on poppy
3314
Opioid receptors on the surfaces of brain cells are involved in pleasure, pain, addiction, depression, psychosis, and other conditions. Raymond Stevens, The Scripps Research Institute View MediaProtein involved in cell division from Mycoplasma pneumoniae
2377
Model of a protein involved in cell division from Mycoplasma pneumoniae. This model, based on X-ray crystallography, revealed a structural domain not seen before. Berkeley Structural Genomics Center, PSI View MediaStructure of amyloid-forming prion protein
3542
This structure from an amyloid-forming prion protein shows one way beta sheets can stack. Douglas Fowler, University of Washington View MediaX-ray co-crystal structure of Src kinase bound to a DNA-templated macrocycle inhibitor 2
3414
X-ray co-crystal structure of Src kinase bound to a DNA-templated macrocycle inhibitor. Markus A. Seeliger, Stony Brook University Medical School and David R. Liu, Harvard University View MediaSeeing signaling protein activation in cells 03
2453
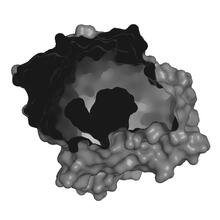
Cdc42, a member of the Rho family of small guanosine triphosphatase (GTPase) proteins, regulates multiple cell functions, including motility, proliferation, apoptosis, and cell morphology. Klaus Hahn, University of North Carolina, Chapel Hill Medical School View MediaAntitoxin GhoS (Illustration 1)
3427
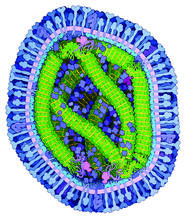
Structure of the bacterial antitoxin protein GhoS. GhoS inhibits the production of a bacterial toxin, GhoT, which can contribute to antibiotic resistance. Rebecca Page and Wolfgang Peti, Brown University and Thomas K. Wood, Pennsylvania State University View MediaCryo-electron microscopy of the dengue virus showing protective membrane and membrane proteins
3748
Dengue virus is a mosquito-borne illness that infects millions of people in the tropics and subtropics each year. Like many viruses, dengue is enclosed by a protective membrane. Hong Zhou, UCLA View MediaX-ray co-crystal structure of Src kinase bound to a DNA-templated macrocycle inhibitor 6
3418
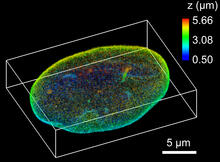
X-ray co-crystal structure of Src kinase bound to a DNA-templated macrocycle inhibitor. Markus A. Seeliger, Stony Brook University Medical School and David R. Liu, Harvard University View MediaNuclear Lamina – Three Views
6573
Three views of the entire nuclear lamina of a HeLa cell produced by tilted light sheet 3D single-molecule super-resolution imaging using a platform termed TILT3D. Anna-Karin Gustavsson, Ph.D. View MediaPig trypsin (1)
2400
A crystal of porcine trypsin protein created for X-ray crystallography, which can reveal detailed, three-dimensional protein structures. Alex McPherson, University of California, Irvine View MediaCryo-electron microscopy revealing the "wasabi receptor"
3747
The TRPA1 protein is responsible for the burn you feel when you taste a bite of sushi topped with wasabi. Jean-Paul Armache, UCSF View MediaProtein from Methanobacterium thermoautotrophicam
2374
A knotted protein from an archaebacterium called Methanobacterium thermoautotrophicam. This organism breaks down waste products and produces methane gas. Midwest Center For Structural Genomics, PSI View MediaAtomic-level structure of the HIV capsid
6601
This animation shows atoms of the HIV capsid, the shell that encloses the virus's genetic material. Juan R. Perilla and the Theoretical and Computational Biophysics Group, University of Illinois at Urbana-Champaign View MediaMicrotubule dynamics in real time
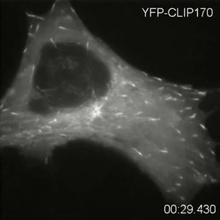
2784
Cytoplasmic linker protein (CLIP)-170 is a microtubule plus-end-tracking protein that regulates microtubule dynamics and links microtubule ends to different intracellular structures. Gary Borisy, Marine Biology Laboratory View MediaDengue virus membrane protein structure
3758
Dengue virus is a mosquito-borne illness that infects millions of people in the tropics and subtropics each year. Like many viruses, dengue is enclosed by a protective membrane. Hong Zhou, UCLA View MediaHen egg lysozyme (1)
2396
Crystals of hen egg lysozyme protein created for X-ray crystallography, which can reveal detailed, three-dimensional protein structures. Alex McPherson, University of California, Irvine View MediaEarly life of a protein
2740
This illustration represents the early life of a protein—specifically, apomyoglobin—as it is synthesized by a ribosome and emerges from the ribosomal tunnel, which contains the newly formed protein's Silvia Cavagnero, University of Wisconsin, Madison View MediaKinesin moves cellular cargo
3491
A protein called kinesin (blue) is in charge of moving cargo around inside cells and helping them divide. Charles Sindelar, Yale University View MediaBovine milk alpha-lactalbumin (1)
2397
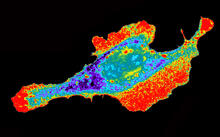
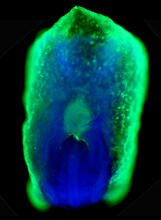
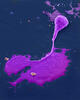
A crystal of bovine milk alpha-lactalbumin protein created for X-ray crystallography, which can reveal detailed, three-dimensional protein structures. Alex McPherson, University of California, Irvine View MediaMouse embryo showing Smad4 protein
2607
This eerily glowing blob isn't an alien or a creature from the deep sea--it's a mouse embryo just eight and a half days old. The green shell and core show a protein called Smad4. Kenneth Zaret, Fox Chase Cancer Center View MediaNuclear Lamina
6572
The 3D single-molecule super-resolution reconstruction of the entire nuclear lamina in a HeLa cell was acquired using the TILT3D platform. Anna-Karin Gustavsson, Ph.D. View MediaATP Synthase
6353
Atomic model of the membrane region of the mitochondrial ATP synthase built into a cryo-EM map at 3.6 Å resolution. ATP synthase is the primary producer of ATP in aerobic cells. Bridget Carragher, <a href="http://nramm.nysbc.org/">NRAMM National Resource for Automated Molecular Microscopy</a> View MediaCatalase diversity
7003
Catalases are some of the most efficient enzymes found in cells. Amy Wu and Christine Zardecki, RCSB Protein Data Bank. View MediaFlower-forming cells in a small plant related to cabbage (Arabidopsis)
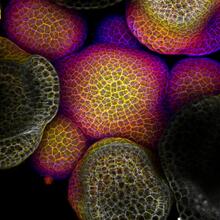
3606
In plants, as in animals, stem cells can transform into a variety of different cell types. The stem cells at the growing tip of this Arabidopsis plant will soon become flowers. Arun Sampathkumar and Elliot Meyerowitz, California Institute of Technology View MediaOptic nerve astrocytes
5852
Astrocytes in the cross section of a human optic nerve head Tom Deerinck and Keunyoung (“Christine”) Kim, NCMIR View MediaDisease-susceptible Arabidopsis leaf
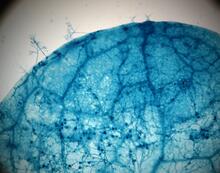
2782
This is a magnified view of an Arabidopsis thaliana leaf after several days of infection with the pathogen Hyaloperonospora arabidopsidis. Jeff Dangl, University of North Carolina, Chapel Hill View MediaDiversity oriented synthesis: generating skeletal diversity using folding processes
3327
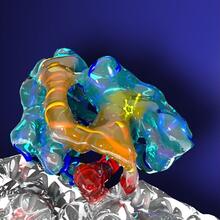
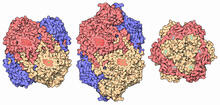
This 1 1/2-minute video animation was produced for chemical biologist Stuart Schreiber's lab page. The animation shows how diverse chemical structures can be produced in the lab. Eric Keller View MediaIntasome
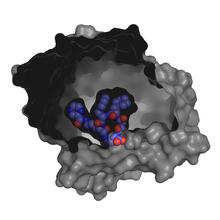
6346
Salk researchers captured the structure of a protein complex called an intasome (center) that lets viruses similar to HIV establish permanent infection in their hosts. National Resource for Automated Molecular Microscopy http://nramm.nysbc.org/nramm-images/ Source: Bridget Carragher View MediaProtein crystals
1060
Structural biologists create crystals of proteins, shown here, as a first step in a process called X-ray crystallography, which can reveal detailed, three-dimensional protein structures. Alex McPherson, University of California, Irvine View MediaHen egg lysozyme (2)
2406
A crystal of hen egg lysozyme protein created for X-ray crystallography, which can reveal detailed, three-dimensional protein structures. Alex McPherson, University of California, Irvine View MediaX-ray co-crystal structure of Src kinase bound to a DNA-templated macrocycle inhibitor 3
3415
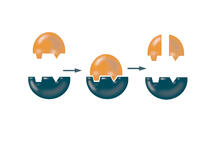
X-ray co-crystal structure of Src kinase bound to a DNA-templated macrocycle inhibitor. Markus A. Seeliger, Stony Brook University Medical School and David R. Liu, Harvard University View MediaHsp33 figure 1
3354
Featured in the March 15, 2012 issue of Biomedical Beat. Related to Hsp33 Figure 2, image 3355. Ursula Jakob and Dana Reichmann, University of Michigan View MediaEnzymes convert subtrates into products
2521
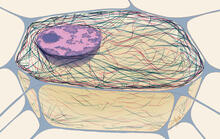
Enzymes convert substrates into products very quickly. See image 2522 for a labeled version of this illustration. Featured in The Chemistry of Health. Crabtree + Company View MediaCytoskeleton
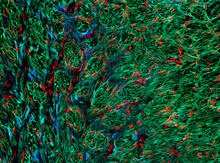
1272
The three fibers of the cytoskeleton--microtubules in blue, intermediate filaments in red, and actin in green--play countless roles in the cell. Judith Stoffer View MediaBrains of sleep-deprived and well-rested fruit flies
3490
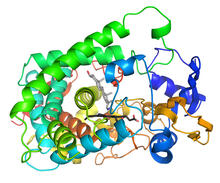
On top, the brain of a sleep-deprived fly glows orange because of Bruchpilot, a communication protein between brain cells. These bright orange brain areas are associated with learning. Chiara Cirelli, University of Wisconsin-Madison View MediaCytochrome structure with anticancer drug
3326
This image shows the structure of the CYP17A1 enzyme (ribbons colored from blue N-terminus to red C-terminus), with the associated heme colored black. Emily Scott, University of Kansas View MediaMeasles virus
6995
A cross section of the measles virus in which six proteins work together to infect cells. The measles virus is extremely infectious; 9 out of 10 people exposed will contract the disease. Amy Wu and Christine Zardecki, RCSB Protein Data Bank. View MediaCryo-EM reveals how the HIV capsid attaches to a human protein to evade immune detection
3755
The illustration shows the capsid of human immunodeficiency virus (HIV) whose molecular features were resolved with cryo-electron microscopy (cryo-EM). Juan R. Perilla, University of Illinois at Urbana-Champaign View MediaBacterial glucose isomerase
2409
A crystal of bacterial glucose isomerase protein created for X-ray crystallography, which can reveal detailed, three-dimensional protein structures. Alex McPherson, University of California, Irvine View MediaRhodopsin bound to visual arrestin
6768
Rhodopsin is a pigment in the rod cells of the retina (back of the eye). It is extremely light-sensitive, supporting vision in low-light conditions. Protein Data Bank. View Media