Switch to List View
Image and Video Gallery
This is a searchable collection of scientific photos, illustrations, and videos. The images and videos in this gallery are licensed under Creative Commons Attribution Non-Commercial ShareAlike 3.0. This license lets you remix, tweak, and build upon this work non-commercially, as long as you credit and license your new creations under identical terms.
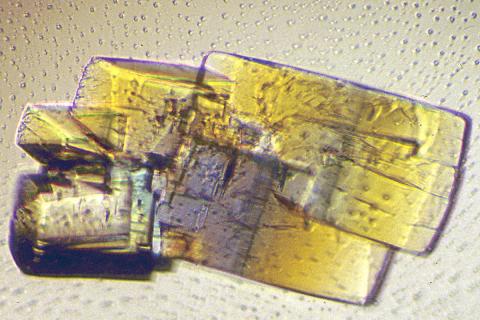
2402: RNase A (2)
2402: RNase A (2)
A crystal of RNase A protein created for X-ray crystallography, which can reveal detailed, three-dimensional protein structures.
Alex McPherson, University of California, Irvine
View Media
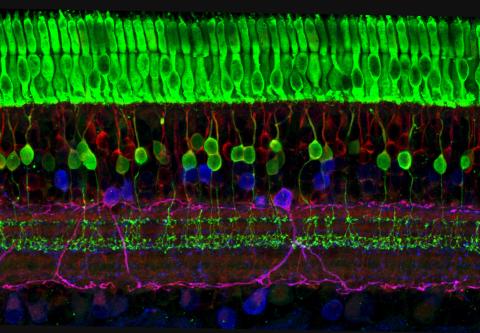
3635: The eye uses many layers of nerve cells to convert light into sight
3635: The eye uses many layers of nerve cells to convert light into sight
This image captures the many layers of nerve cells in the retina. The top layer (green) is made up of cells called photoreceptors that convert light into electrical signals to relay to the brain. The two best-known types of photoreceptor cells are rod- and cone-shaped. Rods help us see under low-light conditions but can't help us distinguish colors. Cones don't function well in the dark but allow us to see vibrant colors in daylight.
This image was part of the Life: Magnified exhibit that ran from June 3, 2014, to January 21, 2015, at Dulles International Airport.
This image was part of the Life: Magnified exhibit that ran from June 3, 2014, to January 21, 2015, at Dulles International Airport.
Wei Li, National Eye Institute, National Institutes of Health
View Media
3583: Bee venom toxin destroying a cell
3583: Bee venom toxin destroying a cell
This video condenses 6.5 minutes into less than a minute to show how the toxin in bee venom, called melittin, destroys an animal or bacterial cell. What looks like a red balloon is an artificial cell filled with red dye. Melittin molecules are colored green and float on the cell's surface like twigs on a pond. As melittin accumulates on the cell's membrane, the membrane expands to accommodate it. In the video, the membrane stretches into a column on the left. When melittin levels reach a critical threshold, countless pinhole leaks burst open in the membrane. The cell's vital fluids (red dye in the video) leak out through these pores. Within minutes, the cell collapses.
Huey Huang, Rice University
View Media
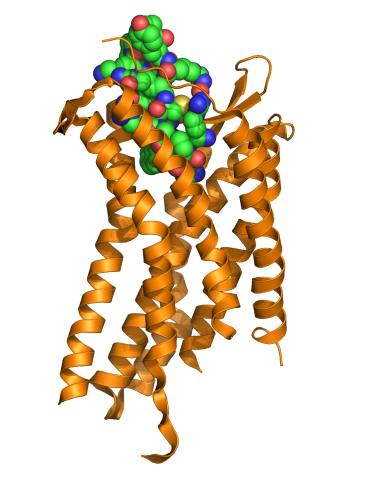
3365: Chemokine CXCR4 receptor
3365: Chemokine CXCR4 receptor
The receptor is shown bound to a small molecule peptide called CVX15.
Raymond Stevens, The Scripps Research Institute
View Media
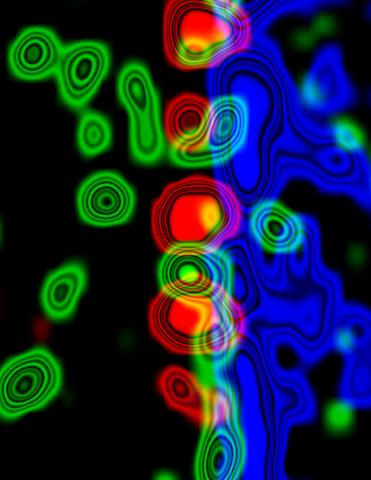
3734: Molecular interactions at the astrocyte nuclear membrane
3734: Molecular interactions at the astrocyte nuclear membrane
These ripples of color represent the outer membrane of the nucleus inside an astrocyte, a star-shaped cell inside the brain. Some proteins (green) act as keys to unlock other proteins (red) that form gates to let small molecules in and out of the nucleus (blue). Visualizing these different cell components at the boundary of the astrocyte nucleus enables researchers to study the molecular and physiological basis of neurological disorders, such as hydrocephalus, a condition in which too much fluid accumulates in the brain, and scar formation in brain tissue leading to abnormal neuronal activity affecting learning and memory. Scientists have now identified a pathway may be common to many of these brain diseases and begun to further examine it to find ways to treat certain brain diseases and injuries.
Katerina Akassoglou, Gladstone Institute for Neurological Disease & UCSF
View Media
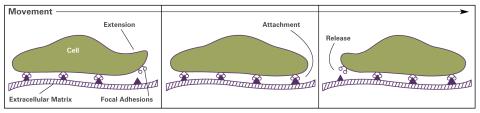
2503: Focal adhesions (with labels)
2503: Focal adhesions (with labels)
Cells walk along body surfaces via tiny "feet," called focal adhesions, that connect with the extracellular matrix. See image 2502 for an unlabeled version of this illustration.
Crabtree + Company
View Media
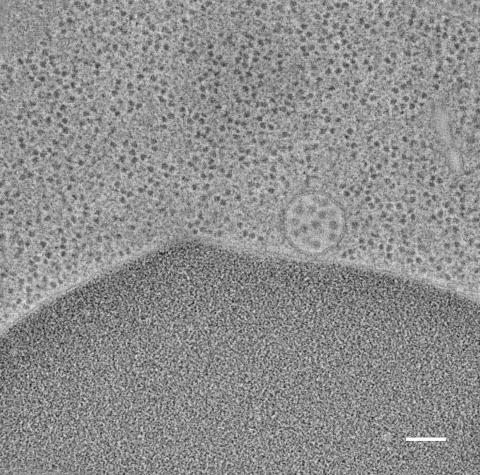
5768: Multivesicular bodies containing intralumenal vesicles assemble at the vacuole 2
5768: Multivesicular bodies containing intralumenal vesicles assemble at the vacuole 2
Collecting and transporting cellular waste and sorting it into recylable and nonrecylable pieces is a complex business in the cell. One key player in that process is the endosome, which helps collect, sort and transport worn-out or leftover proteins with the help of a protein assembly called the endosomal sorting complexes for transport (or ESCRT for short). These complexes help package proteins marked for breakdown into intralumenal vesicles, which, in turn, are enclosed in multivesicular bodies for transport to the places where the proteins are recycled or dumped. In this image, a multivesicular body (the round structure slightly to the right of center) contain tiny intralumenal vesicles (with a diameter of only 25 nanometers; the round specks inside the larger round structure) adjacent to the cell's vacuole (below the multivesicular body, shown in darker and more uniform gray).
Scientists working with baker's yeast (Saccharomyces cerevisiae) study the budding inward of the limiting membrane (green lines on top of the yellow lines) into the intralumenal vesicles. This tomogram was shot with a Tecnai F-20 high-energy electron microscope, at 29,000x magnification, with a 0.7-nm pixel, ~4-nm resolution.
To learn more about endosomes, see the Biomedical Beat blog post The Cell’s Mailroom. Related to a color-enhanced version 5767 and image 5769.
Scientists working with baker's yeast (Saccharomyces cerevisiae) study the budding inward of the limiting membrane (green lines on top of the yellow lines) into the intralumenal vesicles. This tomogram was shot with a Tecnai F-20 high-energy electron microscope, at 29,000x magnification, with a 0.7-nm pixel, ~4-nm resolution.
To learn more about endosomes, see the Biomedical Beat blog post The Cell’s Mailroom. Related to a color-enhanced version 5767 and image 5769.
Matthew West and Greg Odorizzi, University of Colorado
View Media
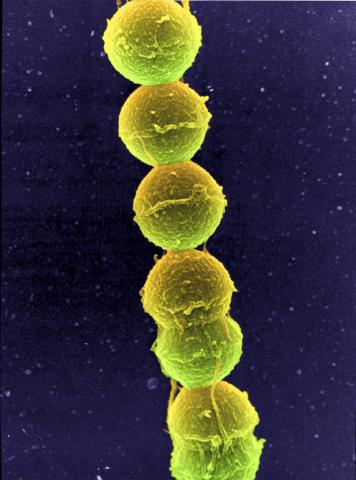
1157: Streptococcus bacteria
1157: Streptococcus bacteria
Image of Streptococcus, a type (genus) of spherical bacteria that can colonize the throat and back of the mouth. Stroptococci often occur in pairs or in chains, as shown here.
Tina Weatherby Carvalho, University of Hawaii at Manoa
View Media

2593: Precise development in the fruit fly embryo
2593: Precise development in the fruit fly embryo
This 2-hour-old fly embryo already has a blueprint for its formation, and the process for following it is so precise that the difference of just a few key molecules can change the plans. Here, blue marks a high concentration of Bicoid, a key signaling protein that directs the formation of the fly's head. It also regulates another important protein, Hunchback (green), that further maps the head and thorax structures and partitions the embryo in half (red is DNA). The yellow dots overlaying the embryo plot the concentration of Bicoid versus Hunchback proteins within each nucleus. The image illustrates the precision with which an embryo interprets and locates its halfway boundary, approaching limits set by simple physical principles. This image was a finalist in the 2008 Drosophila Image Award.
Thomas Gregor, Princeton University
View Media
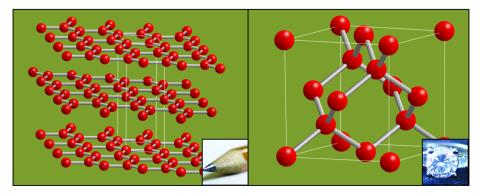
2507: Carbon building blocks (with examples)
2507: Carbon building blocks (with examples)
The arrangement of identical molecular components can make a dramatic difference. For example, carbon atoms can be arranged into dull graphite (left) or sparkly diamonds (right). See image 2506 for an illustration without examples.
Crabtree + Company
View Media
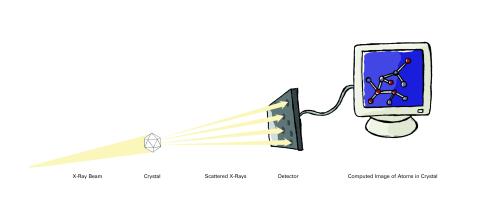
2512: X-ray crystallography (with labels)
2512: X-ray crystallography (with labels)
X-ray crystallography allows researchers to see structures too small to be seen by even the most powerful microscopes. To visualize the arrangement of atoms within molecules, researchers can use the diffraction patterns obtained by passing X-ray beams through crystals of the molecule. This is a common way for solving the structures of proteins. See image 2511 for an unlabeled version of this illustration. Featured in The Structures of Life.
Crabtree + Company
View Media
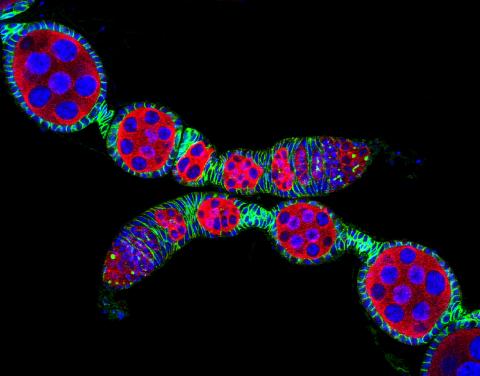
5772: Confocal microscopy image of two Drosophila ovarioles
5772: Confocal microscopy image of two Drosophila ovarioles
Ovarioles in female insects are tubes in which egg cells (called oocytes) form at one end and complete their development as they reach the other end of the tube. This image, taken with a confocal microscope, shows ovarioles in a very popular lab animal, the fruit fly Drosophila. The basic structure of ovarioles supports very rapid egg production, with some insects (like termites) producing several thousand eggs per day. Each insect ovary typically contains four to eight ovarioles, but this number varies widely depending on the insect species.
Scientists use insect ovarioles, for example, to study the basic processes that help various insects, including those that cause disease (like some mosquitos and biting flies), reproduce very quickly.
Scientists use insect ovarioles, for example, to study the basic processes that help various insects, including those that cause disease (like some mosquitos and biting flies), reproduce very quickly.
2004 Olympus BioScapes Competition
View Media
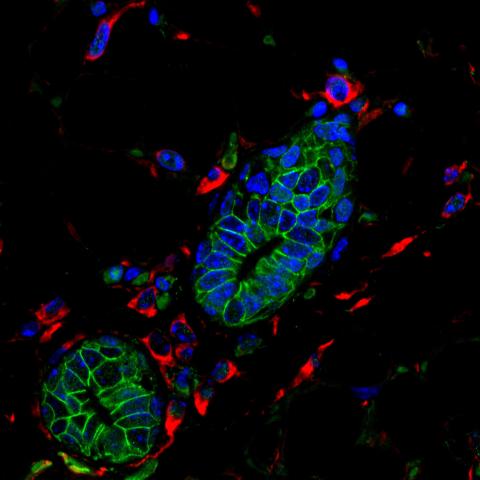
3432: Mouse mammary cells lacking anti-cancer protein
3432: Mouse mammary cells lacking anti-cancer protein
Shortly after a pregnant woman gives birth, her breasts start to secrete milk. This process is triggered by hormonal and genetic cues, including the protein Elf5. Scientists discovered that Elf5 also has another job--it staves off cancer. Early in the development of breast cancer, human breast cells often lose Elf5 proteins. Cells without Elf5 change shape and spread readily--properties associated with metastasis. This image shows cells in the mouse mammary gland that are lacking Elf5, leading to the overproduction of other proteins (red) that increase the likelihood of metastasis.
Nature Cell Biology, November 2012, Volume 14 No 11 pp1113-1231
View Media
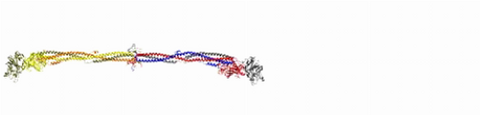
2450: Blood clots show their flex
2450: Blood clots show their flex
Blood clots stop bleeding, but they also can cause heart attacks and strokes. A team led by computational biophysicist Klaus Schulten of the University of Illinois at Urbana-Champaign has revealed how a blood protein can give clots their lifesaving and life-threatening abilities. The researchers combined experimental and computational methods to animate fibrinogen, a protein that forms the elastic fibers that enable clots to withstand the force of blood pressure. This simulation shows that the protein, through a series of events, stretches up to three times its length. Adjusting this elasticity could improve how we manage healthful and harmful clots. NIH's National Center for Research Resources also supported this work. Featured in the March 19, 2008, issue of Biomedical Beat.
Eric Lee, University of Illinois at Urbana-Champaign
View Media
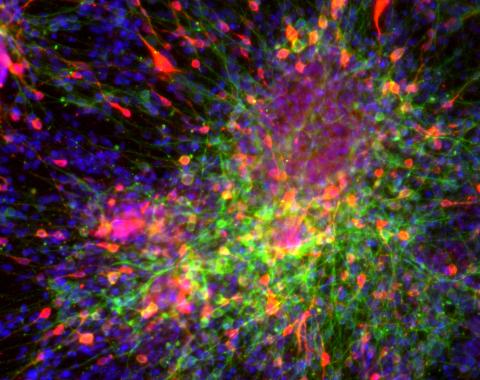
3271: Dopaminergic neurons derived from mouse embryonic stem cells
3271: Dopaminergic neurons derived from mouse embryonic stem cells
These neurons are derived from mouse embryonic stem cells. Red shows cells making a protein called TH that is characteristic of the neurons that degenerate in Parkinson's disease. Green indicates a protein that's found in all neurons. Blue indicates the nuclei of all cells. Studying dopaminergic neurons can help researchers understand the origins of Parkinson's disease and could be used to screen potential new drugs. Image and caption information courtesy of the California Institute for Regenerative Medicine. Related to images 3270 and 3285.
Yaping Sun, lab of Su Guo, University of California, San Francisco, via CIRM
View Media
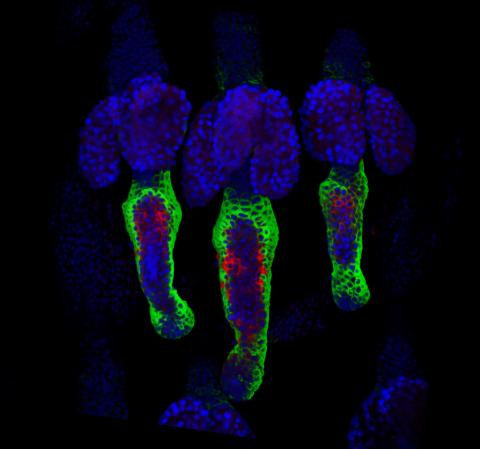
3497: Wound healing in process
3497: Wound healing in process
Wound healing requires the action of stem cells. In mice that lack the Sept2/ARTS gene, stem cells involved in wound healing live longer and wounds heal faster and more thoroughly than in normal mice. This confocal microscopy image from a mouse lacking the Sept2/ARTS gene shows a tail wound in the process of healing. See more information in the article in Science.
Related to images 3498 and 3500.
Related to images 3498 and 3500.
Hermann Steller, Rockefeller University
View Media

6886: Neutrophil-like cells migrating in a microfluidic chip
6886: Neutrophil-like cells migrating in a microfluidic chip
Neutrophil-like cells (blue) in a microfluidic chip preferentially migrating toward LTB4 over fMLP. A neutrophil is a type of white blood cell that is part of the immune system and helps the body fight infection. Both LTB4 and fMLP are molecules involved in immune response. Microfluidic chips are small devices containing microscopic channels, and they are used in a range of applications, from basic research on cells to pathogen detection. The scale bar in this video is 500μm.
Caroline Jones, University of Texas at Dallas.
View Media
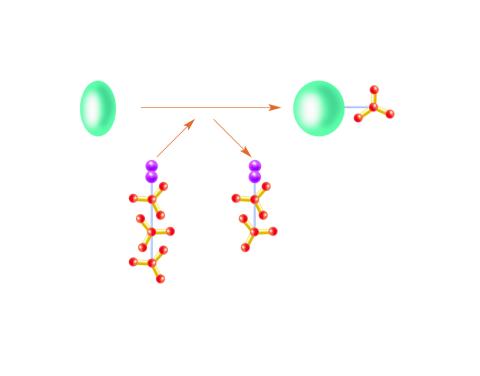
2534: Kinases
2534: Kinases
Kinases are enzymes that add phosphate groups (red-yellow structures) to proteins (green), assigning the proteins a code. In this reaction, an intermediate molecule called ATP (adenosine triphosphate) donates a phosphate group from itself, becoming ADP (adenosine diphosphate). See image 2535 for a labeled version of this illustration. Featured in Medicines By Design.
Crabtree + Company
View Media
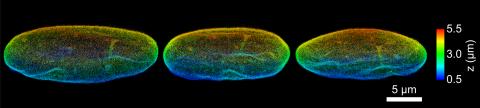
6573: Nuclear Lamina – Three Views
6573: Nuclear Lamina – Three Views
Three views of the entire nuclear lamina of a HeLa cell produced by tilted light sheet 3D single-molecule super-resolution imaging using a platform termed TILT3D.
See 6572 for a 3D view of this structure.
See 6572 for a 3D view of this structure.
Anna-Karin Gustavsson, Ph.D.
View Media
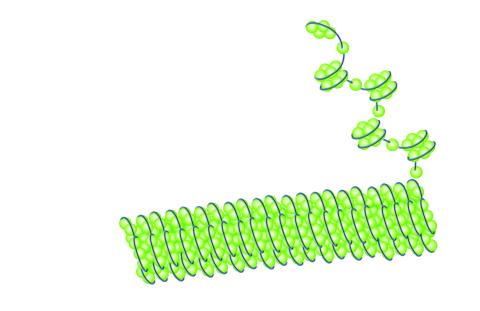
2560: Histones in chromatin
2560: Histones in chromatin
Histone proteins loop together with double-stranded DNA to form a structure that resembles beads on a string. See image 2561 for a labeled version of this illustration. Featured in The New Genetics.
Crabtree + Company
View Media
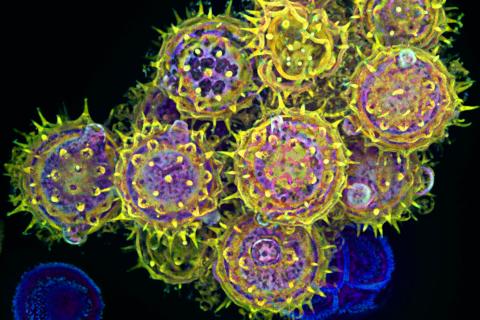
3609: Pollen grains: male germ cells in plants and a cause of seasonal allergies
3609: Pollen grains: male germ cells in plants and a cause of seasonal allergies
Those of us who get sneezy and itchy-eyed every spring or fall may have pollen grains, like those shown here, to blame. Pollen grains are the male germ cells of plants, released to fertilize the corresponding female plant parts. When they are instead inhaled into human nasal passages, they can trigger allergies.
This image was part of the Life: Magnified exhibit that ran from June 3, 2014, to January 21, 2015, at Dulles International Airport.
This image was part of the Life: Magnified exhibit that ran from June 3, 2014, to January 21, 2015, at Dulles International Airport.
Edna, Gil, and Amit Cukierman, Fox Chase Cancer Center, Philadelphia, Pa.
View Media
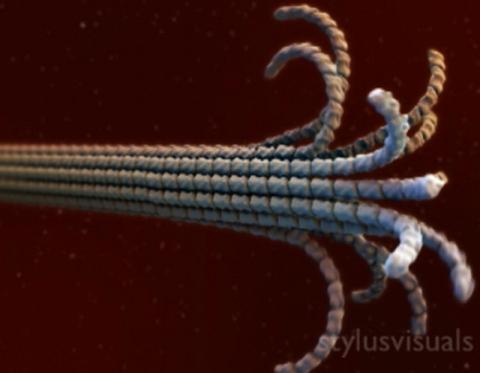
2321: Microtubule breakdown
2321: Microtubule breakdown
Like a building supported by a steel frame, a cell contains its own sturdy internal scaffolding made up of proteins, including microtubules. Researchers studying snapshots of microtubules have proposed a model for how these structural elements shorten and lengthen, allowing a cell to move, divide, or change shape. This picture shows an intermediate step in the model: Smaller building blocks called tubulins peel back from the microtubule in thin strips. Knowing the operations of the internal scaffolding will enhance our basic understanding of cellular processes.
Eva Nogales, University of California, Berkeley
View Media
3449: Calcium uptake during ATP production in mitochondria
3449: Calcium uptake during ATP production in mitochondria
Living primary mouse embryonic fibroblasts. Mitochondria (green) stained with the mitochondrial membrane potential indicator, rhodamine 123. Nuclei (blue) are stained with DAPI. Caption from a November 26, 2012 news release from U Penn (Penn Medicine).
Lili Guo, Perelman School of Medicine, University of Pennsylvania
View Media
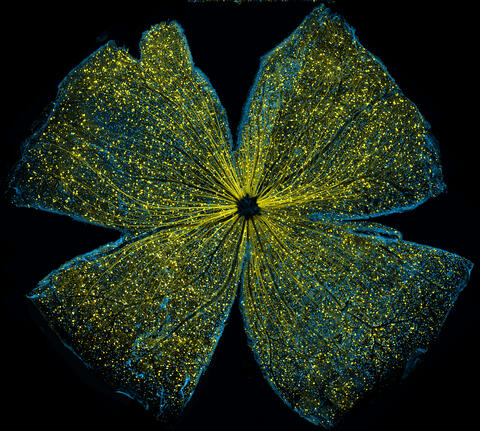
5793: Mouse retina
5793: Mouse retina
What looks like the gossamer wings of a butterfly is actually the retina of a mouse, delicately snipped to lay flat and sparkling with fluorescent molecules. The image is from a research project investigating the promise of gene therapy for glaucoma. It was created at an NIGMS-funded advanced microscopy facility that develops technology for imaging across many scales, from whole organisms to cells to individual molecules.
The ability to obtain high-resolution imaging of tissue as large as whole mouse retinas was made possible by a technique called large-scale mosaic confocal microscopy, which was pioneered by the NIGMS-funded National Center for Microscopy and Imaging Research. The technique is similar to Google Earth in that it computationally stitches together many small, high-resolution images.
The ability to obtain high-resolution imaging of tissue as large as whole mouse retinas was made possible by a technique called large-scale mosaic confocal microscopy, which was pioneered by the NIGMS-funded National Center for Microscopy and Imaging Research. The technique is similar to Google Earth in that it computationally stitches together many small, high-resolution images.
Tom Deerinck and Keunyoung (“Christine”) Kim, NCMIR
View Media
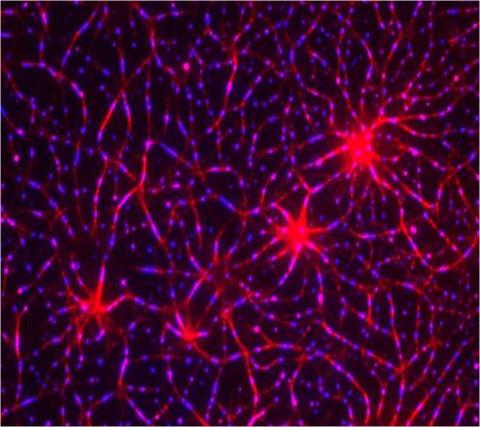
3787: In vitro assembly of a cell-signaling pathway
3787: In vitro assembly of a cell-signaling pathway
T cells are white blood cells that are important in defending the body against bacteria, viruses and other pathogens. Each T cell carries proteins, called T-cell receptors, on its surface that are activated when they come in contact with an invader. This activation sets in motion a cascade of biochemical changes inside the T cell to mount a defense against the invasion. Scientists have been interested for some time what happens after a T-cell receptor is activated. One obstacle has been to study how this signaling cascade, or pathway, proceeds inside T cells.
In this image, researchers have created a T-cell receptor pathway consisting of 12 proteins outside the cell on an artificial membrane. The image shows two key steps during the signaling process: clustering of a protein called linker for activation of T cells (LAT) (blue) and polymerization of the cytoskeleton protein actin (red). The findings show that the T-cell receptor signaling proteins self-organize into separate physical and biochemical compartments. This new system of studying molecular pathways outside the cells will enable scientists to better understand how the immune system combats microbes or other agents that cause infection.
To learn more how researchers assembled this T-cell receptor pathway, see this press release from HHMI's Marine Biological Laboratory Whitman Center. Related to video 3786.
In this image, researchers have created a T-cell receptor pathway consisting of 12 proteins outside the cell on an artificial membrane. The image shows two key steps during the signaling process: clustering of a protein called linker for activation of T cells (LAT) (blue) and polymerization of the cytoskeleton protein actin (red). The findings show that the T-cell receptor signaling proteins self-organize into separate physical and biochemical compartments. This new system of studying molecular pathways outside the cells will enable scientists to better understand how the immune system combats microbes or other agents that cause infection.
To learn more how researchers assembled this T-cell receptor pathway, see this press release from HHMI's Marine Biological Laboratory Whitman Center. Related to video 3786.
Xiaolei Su, HHMI Whitman Center of the Marine Biological Laboratory
View Media
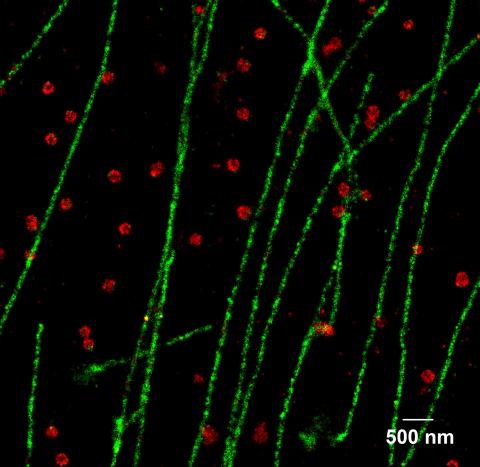
2325: Multicolor STORM
2325: Multicolor STORM
In 2006, scientists developed an optical microscopy technique enabling them to clearly see individual molecules within cells. In 2007, they took the technique, abbreviated STORM, a step further. They identified multicolored probes that let them peer into cells and clearly see multiple cellular components at the same time, such as these microtubules (green) and small hollows called clathrin-coated pits (red). Unlike conventional methods, the multicolor STORM technique produces a crisp and high resolution picture. A sharper view of how cellular components interact will likely help scientists answer some longstanding questions about cell biology.
Xiaowei Zhuang, Harvard University
View Media
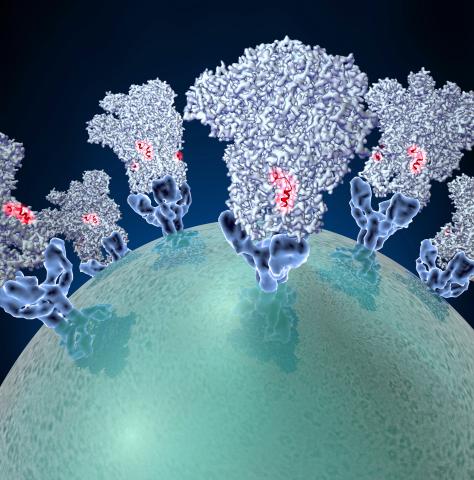
3753: Coronavirus spike protein structure
3753: Coronavirus spike protein structure
Coronaviruses are enveloped viruses responsible for 30 percent of mild respiratory infections and atypical deadly pneumonia in humans worldwide. These deadly pneumonia include those caused by infections with severe acute respiratory syndrome coronavirus (SARS-CoV) and Middle East respiratory syndrome coronavirus (MERS-CoV). The coronavirus spike glycoprotein mediates virus entry into cells and represents an important therapeutic target. The illustration shows a viral membrane decorated with spike glycoproteins; highlighted in red is a potential neutralization site, which is a protein sequence that might be used as a target for vaccines to combat viruses such as MERS-CoV and other coronaviruses.
Melody Campbell, UCSF
View Media
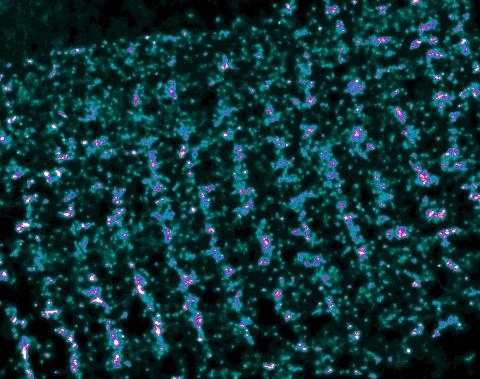
3339: Single-Molecule Imaging
3339: Single-Molecule Imaging
This is a super-resolution light microscope image taken by Hiro Hakozaki and Masa Hoshijima of NCMIR. The image contains highlighted calcium channels in cardiac muscle using a technique called dSTORM. The microscope used in the NCMIR lab was built by Hiro Hakozaki.
Tom Deerinck, NCMIR
View Media
1087: Natcher Building 07
1087: Natcher Building 07
NIGMS staff are located in the Natcher Building on the NIH campus.
Alisa Machalek, National Institute of General Medical Sciences
View Media
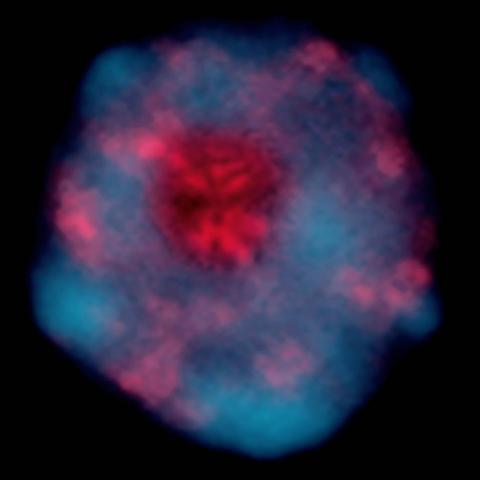
2318: Gene silencing
2318: Gene silencing
Pretty in pink, the enzyme histone deacetylase (HDA6) stands out against a background of blue-tinted DNA in the nucleus of an Arabidopsis plant cell. Here, HDA6 concentrates in the nucleolus (top center), where ribosomal RNA genes reside. The enzyme silences the ribosomal RNA genes from one parent while those from the other parent remain active. This chromosome-specific silencing of ribosomal RNA genes is an unusual phenomenon observed in hybrid plants.
Olga Pontes and Craig Pikaard, Washington University
View Media

2313: Colorful communication
2313: Colorful communication
The marine bacterium Vibrio harveyi glows when near its kind. This luminescence, which results from biochemical reactions, is part of the chemical communication used by the organisms to assess their own population size and distinguish themselves from other types of bacteria. But V. harveyi only light up when part of a large group. This communication, called quorum sensing, speaks for itself here on a lab dish, where more densely packed areas of the bacteria show up blue. Other types of bacteria use quorum sensing to release toxins, trigger disease, and evade the immune system.
Bonnie Bassler, Princeton University
View Media
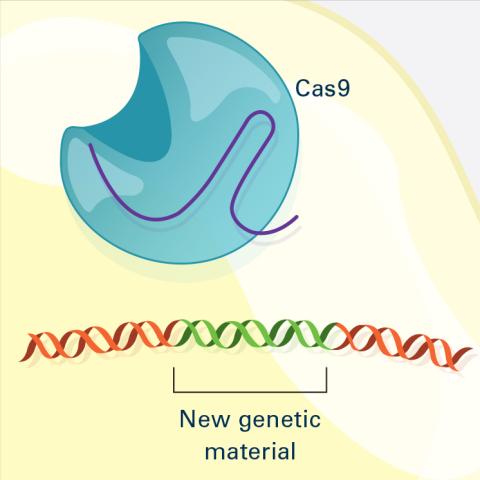
6488: CRISPR Illustration Frame 4
6488: CRISPR Illustration Frame 4
This illustration shows, in simplified terms, how the CRISPR-Cas9 system can be used as a gene-editing tool. The CRISPR system has two components joined together: a finely tuned targeting device (a small strand of RNA programmed to look for a specific DNA sequence) and a strong cutting device (an enzyme called Cas9 that can cut through a double strand of DNA). This frame (4 out of 4) shows a repaired DNA strand with new genetic material that researchers can introduce, which the cell automatically incorporates into the gap when it repairs the broken DNA.
For an explanation and overview of the CRISPR-Cas9 system, see the iBiology video, and find the full CRIPSR illustration here.
For an explanation and overview of the CRISPR-Cas9 system, see the iBiology video, and find the full CRIPSR illustration here.
National Institute of General Medical Sciences.
View Media
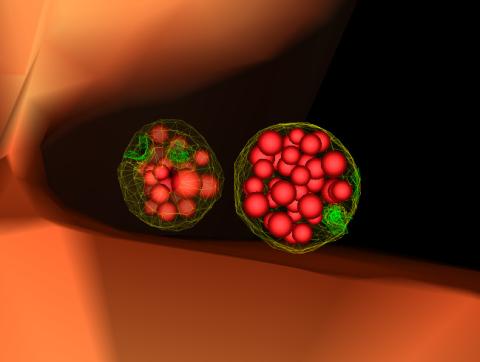
5767: Multivesicular bodies containing intralumenal vesicles assemble at the vacuole 3
5767: Multivesicular bodies containing intralumenal vesicles assemble at the vacuole 3
Collecting and transporting cellular waste and sorting it into recylable and nonrecylable pieces is a complex business in the cell. One key player in that process is the endosome, which helps collect, sort and transport worn-out or leftover proteins with the help of a protein assembly called the endosomal sorting complexes for transport (or ESCRT for short). These complexes help package proteins marked for breakdown into intralumenal vesicles, which, in turn, are enclosed in multivesicular bodies for transport to the places where the proteins are recycled or dumped. In this image, two multivesicular bodies (with yellow membranes) contain tiny intralumenal vesicles (with a diameter of only 25 nanometers; shown in red) adjacent to the cell's vacuole (in orange).
Scientists working with baker's yeast (Saccharomyces cerevisiae) study the budding inward of the limiting membrane (green lines on top of the yellow lines) into the intralumenal vesicles. This tomogram was shot with a Tecnai F-20 high-energy electron microscope, at 29,000x magnification, with a 0.7-nm pixel, ~4-nm resolution.
To learn more about endosomes, see the Biomedical Beat blog post The Cell’s Mailroom. Related to a microscopy photograph 5768 that was used to generate this illustration and a zoomed-out version 5769 of this illustration.
Scientists working with baker's yeast (Saccharomyces cerevisiae) study the budding inward of the limiting membrane (green lines on top of the yellow lines) into the intralumenal vesicles. This tomogram was shot with a Tecnai F-20 high-energy electron microscope, at 29,000x magnification, with a 0.7-nm pixel, ~4-nm resolution.
To learn more about endosomes, see the Biomedical Beat blog post The Cell’s Mailroom. Related to a microscopy photograph 5768 that was used to generate this illustration and a zoomed-out version 5769 of this illustration.
Matthew West and Greg Odorizzi, University of Colorado
View Media

7014: Flagellated bacterial cells
7014: Flagellated bacterial cells
Vibrio fischeri (2 mm in length) is the exclusive symbiotic partner of the Hawaiian bobtail squid, Euprymna scolopes. After this bacterium uses its flagella to swim from the seawater into the light organ of a newly hatched juvenile, it colonizes the host and loses the appendages. This image was taken using a scanning electron microscope.
Margaret J. McFall-Ngai, Carnegie Institution for Science/California Institute of Technology, and Edward G. Ruby, California Institute of Technology.
View Media
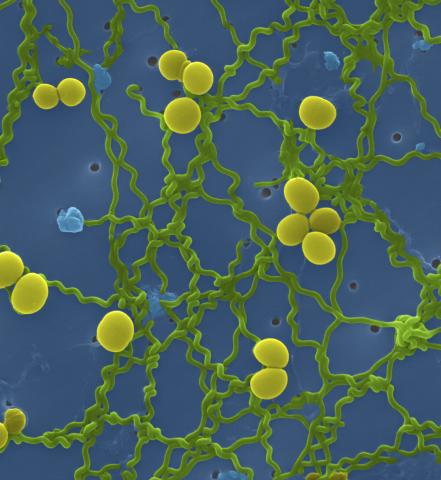
1166: Leptospira bacteria
1166: Leptospira bacteria
Leptospira, shown here in green, is a type (genus) of elongated, spiral-shaped bacteria. Infection can cause Weil's disease, a kind of jaundice, in humans.
Tina Weatherby Carvalho, University of Hawaii at Manoa
View Media
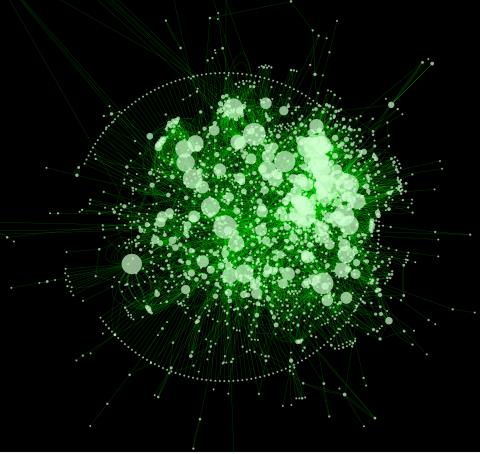
2737: Cytoscape network diagram 1
2737: Cytoscape network diagram 1
Molecular biologists are increasingly relying on bioinformatics software to visualize molecular interaction networks and to integrate these networks with data such as gene expression profiles. Related to 2749.
Keiichiro Ono, Trey Ideker lab, University of California, San Diego
View Media

3728: Quorum-sensing inhibitor limits bacterial growth
3728: Quorum-sensing inhibitor limits bacterial growth
To simulate the consequences of disrupting bacterial cell-to-cell communication, called quorum sensing, in the crypts (small chambers within the colon), the researchers experimented with an inhibitor molecule (i.e., antagonist) to turn off quorum sensing in methicillin-resistant Staphylococcus aureus (MRSA), an antibiotic-resistant strain of bacteria that often causes human infections. In this experiment, a medium promoting bacterial growth flows through experimental chambers mimicking the colon environment. The chambers on the right contained no antagonist. In the left chambers, after being added to the flowing medium, the quorum-sensing-inhibiting molecules quickly spread throughout the crevices, inactivating quorum sensing and reducing colonization. These results suggest a potential strategy for addressing MRSA virulence via inhibitors of bacterial communication. You can read more about this research here.
Minyoung Kevin Kim and Bonnie Bassler, Princeton University
View Media
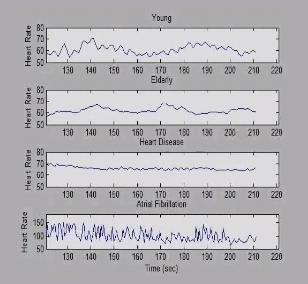
3596: Heart rates time series image
3596: Heart rates time series image
These time series show the heart rates of four different individuals. Automakers use steel scraps to build cars, construction companies repurpose tires to lay running tracks, and now scientists are reusing previously discarded medical data to better understand our complex physiology. Through a website called PhysioNet developed in part by Beth Israel Deaconess Medical Center cardiologist Ary Goldberger, scientists can access complete physiologic recordings, such as heart rate, respiration, brain activity and gait. They then can use free software to analyze the data and find patterns in it. The patterns could ultimately help health care professionals diagnose and treat health conditions like congestive heart failure, sleeping disorders, epilepsy and walking problems. PhysioNet is supported by NIH's National Institute of Biomedical Imaging and Bioengineering as well as by NIGMS.
Madalena Costa and Ary Goldberger, Beth Israel Deaconess Medical Center
View Media
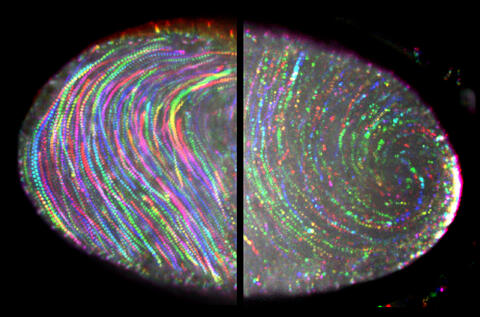
6809: Fruit fly egg ooplasmic streaming
6809: Fruit fly egg ooplasmic streaming
Two fruit fly (Drosophila melanogaster) egg cells, one on each side of the central black line. The colorful swirls show the circular movement of cytoplasm—called ooplasmic streaming—that occurs in late egg cell development in wild-type (right) and mutant (left) oocytes. This image was captured using confocal microscopy.
More information on the research that produced this image can be found in the Journal of Cell Biology paper “Ooplasmic flow cooperates with transport and anchorage in Drosophila oocyte posterior determination” by Lu et al.
More information on the research that produced this image can be found in the Journal of Cell Biology paper “Ooplasmic flow cooperates with transport and anchorage in Drosophila oocyte posterior determination” by Lu et al.
Vladimir I. Gelfand, Feinberg School of Medicine, Northwestern University.
View Media
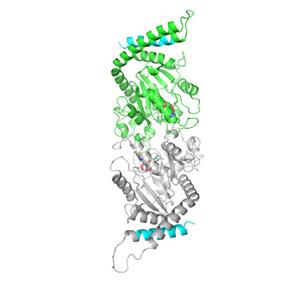
2744: Dynamin structure
2744: Dynamin structure
When a molecule arrives at a cell's outer membrane, the membrane creates a pouch around the molecule that protrudes inward. Directed by a protein called dynamin, the pouch then gets pinched off to form a vesicle that carries the molecule to the right place inside the cell. To better understand how dynamin performs its vital pouch-pinching role, researchers determined its structure. Based on the structure, they proposed that a dynamin "collar" at the pouch's base twists ever tighter until the vesicle pops free. Because cells absorb many drugs through vesicles, the discovery could lead to new drug delivery methods.
Josh Chappie, National Institute of Diabetes and Digestive and Kidney Diseases, NIH
View Media
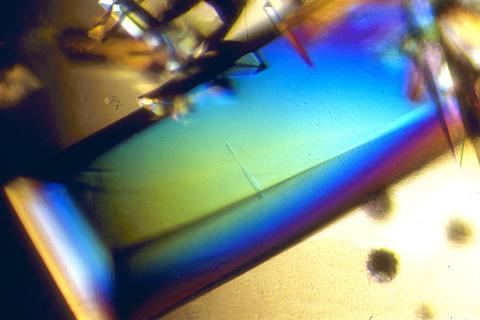
2408: Bovine trypsin
2408: Bovine trypsin
A crystal of bovine trypsin protein created for X-ray crystallography, which can reveal detailed, three-dimensional protein structures.
Alex McPherson, University of California, Irvine
View Media
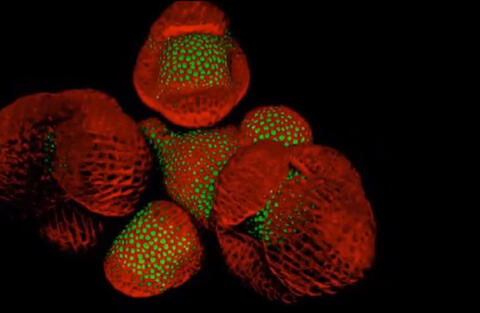
6503: Arabidopsis Thaliana: Flowers Spring to Life
6503: Arabidopsis Thaliana: Flowers Spring to Life
This image capture shows how a single gene, STM, plays a starring role in plant development. This gene acts like a molecular fountain of youth, keeping cells ever-young until it’s time to grow up and commit to making flowers and other plant parts. Because of its ease of use and low cost, Arabidopsis is a favorite model for scientists to learn the basic principles driving tissue growth and regrowth for humans as well as the beautiful plants outside your window. Image captured from video Watch Flowers Spring to Life, featured in the NIH Director's Blog: Watch Flowers Spring to Life.
Nathanaёl Prunet NIH Support: National Institute of General Medical Sciences
View Media
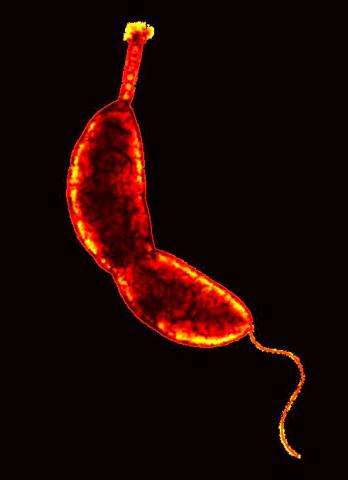
3262: Caulobacter
3262: Caulobacter
A study using Caulobacter crescentus showed that some bacteria use just-in-time processing, much like that used in industrial delivery, to make the glue that allows them to attach to surfaces, an important step in the infection process for many disease-causing bacteria. In the image shown, this freshwater bacterium has a holdfast at the top and a propelling flagellum at the end. From an Indiana University news release.
Yves Brun, Indiana University
View Media
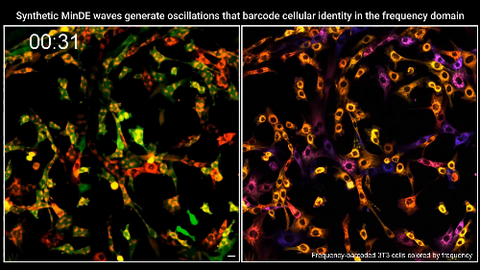
7022: Single-cell “radios” video
7022: Single-cell “radios” video
Individual cells are color-coded based on their identity and signaling activity using a protein circuit technology developed by the Coyle Lab. Just as a radio allows you to listen to an individual frequency, this technology allows researchers to tune into the specific “radio station” of each cell through genetically encoded proteins from a bacterial system called MinDE. The proteins generate an oscillating fluorescent signal that transmits information about cell shape, state, and identity that can be decoded using digital signal processing tools originally designed for telecommunications. The approach allows researchers to look at the dynamics of a single cell in the presence of many other cells.
Related to image 7021.
Related to image 7021.
Scott Coyle, University of Wisconsin-Madison.
View Media
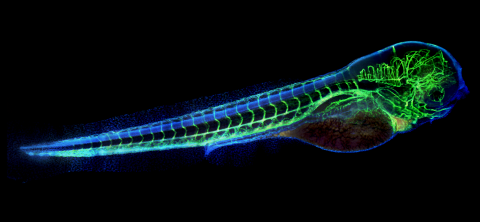
6661: Zebrafish embryo showing vasculature
6661: Zebrafish embryo showing vasculature
A zebrafish embryo. The blue areas are cell bodies, the green lines are blood vessels, and the red glow is blood. This image was created by stitching together five individual images captured with a hyperspectral multipoint confocal fluorescence microscope that was developed at the Eliceiri Lab.
Kevin Eliceiri, University of Wisconsin-Madison.
View Media

2398: RNase A (1)
2398: RNase A (1)
A crystal of RNase A protein created for X-ray crystallography, which can reveal detailed, three-dimensional protein structures.
Alex McPherson, University of California, Irvine
View Media

1336: Life in balance
1336: Life in balance
Mitosis creates cells, and apoptosis kills them. The processes often work together to keep us healthy.
Judith Stoffer
View Media
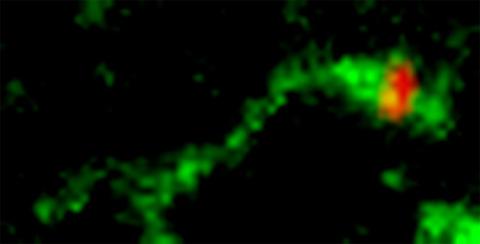
3488: Shiga toxin being sorted inside a cell
3488: Shiga toxin being sorted inside a cell
Shiga toxin (green) is sorted from the endosome into membrane tubules (red), which then pinch off and move to the Golgi apparatus.
Somshuvra Mukhopadhyay, The University of Texas at Austin, and Adam D. Linstedt, Carnegie Mellon University
View Media
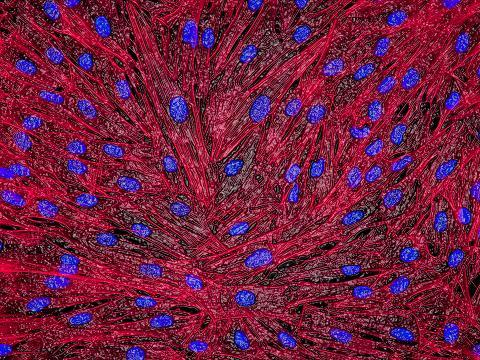
3670: DNA and actin in cultured fibroblast cells
3670: DNA and actin in cultured fibroblast cells
DNA (blue) and actin (red) in cultured fibroblast cells.
Tom Deerinck, National Center for Microscopy and Imaging Research (NCMIR)
View Media