Switch to List View
Image and Video Gallery
This is a searchable collection of scientific photos, illustrations, and videos. The images and videos in this gallery are licensed under Creative Commons Attribution Non-Commercial ShareAlike 3.0. This license lets you remix, tweak, and build upon this work non-commercially, as long as you credit and license your new creations under identical terms.
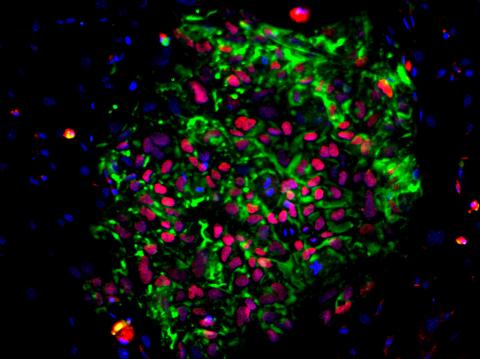
3278: Induced pluripotent stem cells from skin
3278: Induced pluripotent stem cells from skin
These induced pluripotent stem cells (iPS cells) were derived from a woman's skin. Green and red indicate proteins found in reprogrammed cells but not in skin cells (TRA1-62 and NANOG). These cells can then develop into different cell types. Image and caption information courtesy of the California Institute for Regenerative Medicine. Related to image 3279.
Kathrin Plath lab, University of California, Los Angeles, via CIRM
View Media
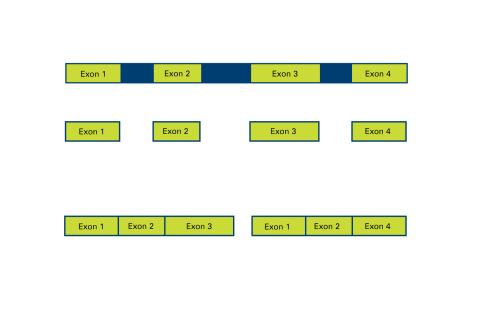
2552: Alternative splicing
2552: Alternative splicing
Arranging exons in different patterns, called alternative splicing, enables cells to make different proteins from a single gene. See image 2553 for a labeled version of this illustration. Featured in The New Genetics.
Crabtree + Company
View Media
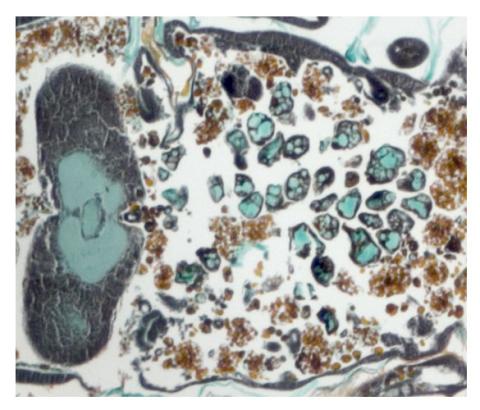
2759: Cross section of a Drosophila melanogaster pupa lacking Draper
2759: Cross section of a Drosophila melanogaster pupa lacking Draper
In the absence of the engulfment receptor Draper, salivary gland cells (light blue) persist in the thorax of a developing Drosophila melanogaster pupa. See image 2758 for a cross section of a normal pupa that does express Draper.
Christina McPhee and Eric Baehrecke, University of Massachusetts Medical School
View Media
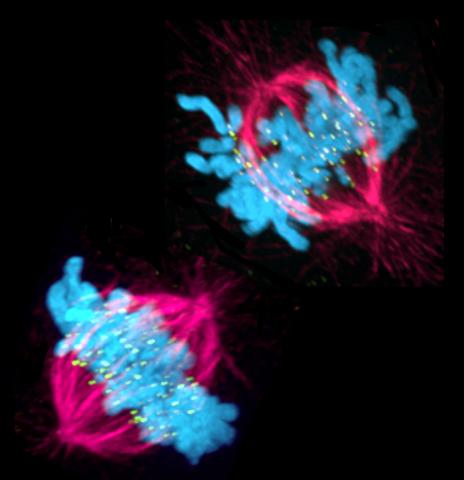
3541: Cell in two stages of division
3541: Cell in two stages of division
This image shows a cell in two stages of division: prometaphase (top) and metaphase (bottom). To form identical daughter cells, chromosome pairs (blue) separate via the attachment of microtubules made up of tubulin proteins (pink) to specialized structures on centromeres (green).
Lilian Kabeche, Dartmouth
View Media
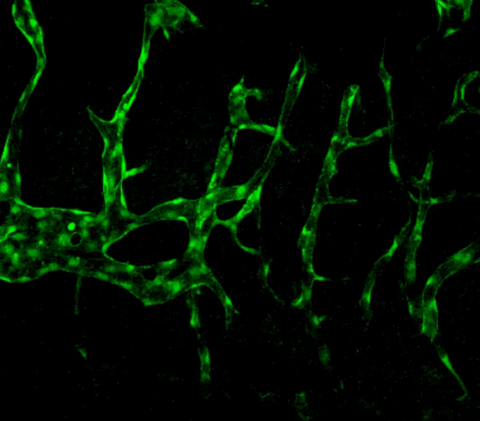
3404: Normal vascular development in frog embryos
3404: Normal vascular development in frog embryos
Hye Ji Cha, University of Texas at Austin
View Media
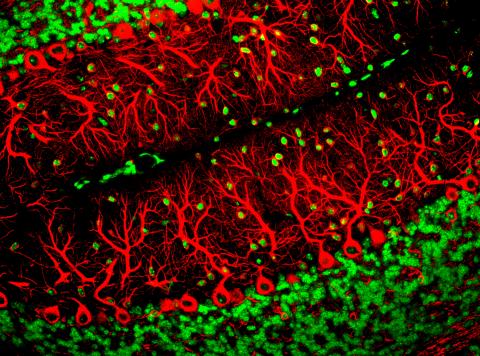
3637: Purkinje cells are one of the main cell types in the brain
3637: Purkinje cells are one of the main cell types in the brain
This image captures Purkinje cells (red), one of the main types of nerve cell found in the brain. These cells have elaborate branching structures called dendrites that receive signals from other nerve cells.
This image was part of the Life: Magnified exhibit that ran from June 3, 2014, to January 21, 2015, at Dulles International Airport.
This image was part of the Life: Magnified exhibit that ran from June 3, 2014, to January 21, 2015, at Dulles International Airport.
Yinghua Ma and Timothy Vartanian, Cornell University, Ithaca, N.Y.
View Media
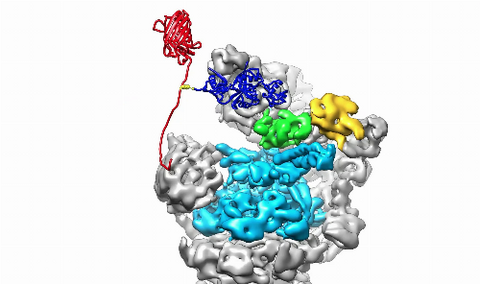
3764: Movie of the 19S proteasome subunit processing a protein substrate
3764: Movie of the 19S proteasome subunit processing a protein substrate
The proteasome is a critical multiprotein complex in the cell that breaks down and recycles proteins that have become damaged or are no longer needed. This movie shows how a protein substrate (red) is bound through its ubiquitin chain (blue) to one of the ubiquitin receptors of the proteasome (Rpn10, yellow). The substrate's flexible engagement region then gets engaged by the AAA+ motor of the proteasome (cyan), which initiates mechanical pulling, unfolding and movement of the protein into the proteasome's interior for cleavage into shorter protein pieces called peptides. During movement of the substrate, its ubiquitin modification gets cleaved off by the deubiquitinase Rpn11 (green), which sits directly above the entrance to the AAA+ motor pore and acts as a gatekeeper to ensure efficient ubiquitin removal, a prerequisite for fast protein breakdown by the 26S proteasome. Related to image 3763.
Andreas Martin, HHMI
View Media
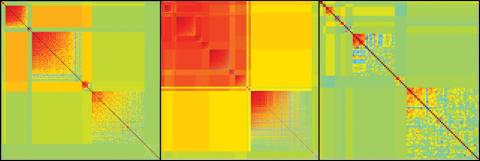
2588: Genetic patchworks
2588: Genetic patchworks
Each point in these colorful patchworks represents the correlation between two sleep-associated genes in fruit flies. Vibrant reds and oranges represent high and intermediate degrees of association between the genes, respectively. Genes in these areas show similar activity patterns in different fly lines. Cool blues represent gene pairs where one partner's activity is high and the other's is low. The green areas show pairs with activities that are not correlated. These quilt-like depictions help illustrate a recent finding that genes act in teams to influence sleep patterns.
Susan Harbison and Trudy Mackay, North Carolina State University
View Media
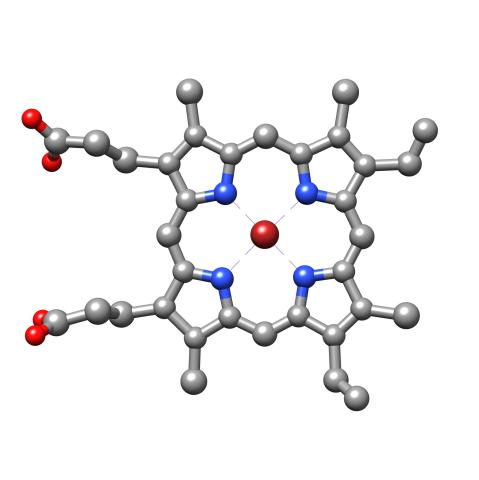
3539: Structure of heme, top view
3539: Structure of heme, top view
Molecular model of the struture of heme. Heme is a small, flat molecule with an iron ion (dark red) at its center. Heme is an essential component of hemoglobin, the protein in blood that carries oxygen throughout our bodies. This image first appeared in the September 2013 issue of Findings Magazine. View side view of heme here 3540.
Rachel Kramer Green, RCSB Protein Data Bank
View Media
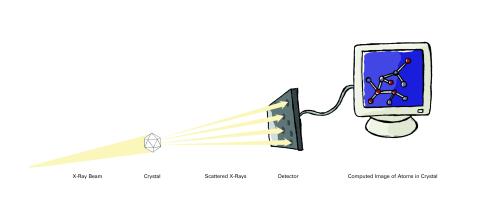
2512: X-ray crystallography (with labels)
2512: X-ray crystallography (with labels)
X-ray crystallography allows researchers to see structures too small to be seen by even the most powerful microscopes. To visualize the arrangement of atoms within molecules, researchers can use the diffraction patterns obtained by passing X-ray beams through crystals of the molecule. This is a common way for solving the structures of proteins. See image 2511 for an unlabeled version of this illustration. Featured in The Structures of Life.
Crabtree + Company
View Media
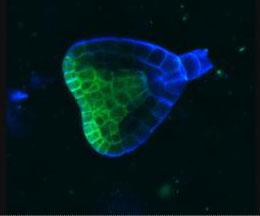
2733: Early development in Arabidopsis
2733: Early development in Arabidopsis
Early on, this Arabidopsis plant embryo picks sides: While one end will form the shoot, the other will take root underground. Short pieces of RNA in the bottom half (blue) make sure that shoot-forming genes are expressed only in the embryo's top half (green), eventually allowing a seedling to emerge with stems and leaves. Like animals, plants follow a carefully orchestrated polarization plan and errors can lead to major developmental defects, such as shoots above and below ground. Because the complex gene networks that coordinate this development in plants and animals share important similarities, studying polarity in Arabidopsis--a model organism--could also help us better understand human development.
Zachery R. Smith, Jeff Long lab at the Salk Institute for Biological Studies
View Media
6522: Fruit fly ovary
6522: Fruit fly ovary
In this image of a stained fruit fly ovary, the ovary is packed with immature eggs (with DNA stained blue). The cytoskeleton (in pink) is a collection of fibers that gives a cell shape and support. The signal-transmitting molecules like STAT (in yellow) are common to reproductive processes in humans. Researchers used this image to show molecular staining and high-resolution imaging techniques to students.
Crystal D. Rogers, Ph.D., University of California, Davis, School of Veterinary Medicine; and Mariano A. Loza-Coll, Ph.D., California State University, Northridge.
View Media
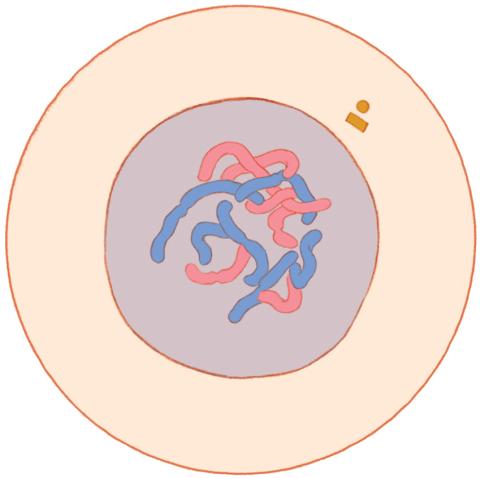
1316: Mitosis - interphase
1316: Mitosis - interphase
A cell in interphase, at the start of mitosis: Chromosomes duplicate, and the copies remain attached to each other. Mitosis is responsible for growth and development, as well as for replacing injured or worn out cells throughout the body. For simplicity, mitosis is illustrated here with only six chromosomes.
Judith Stoffer
View Media
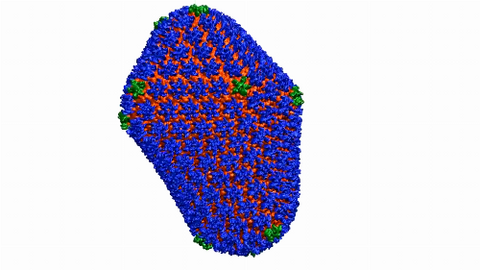
6601: Atomic-level structure of the HIV capsid
6601: Atomic-level structure of the HIV capsid
This animation shows atoms of the HIV capsid, the shell that encloses the virus's genetic material. Scientists determined the exact structure of the capsid using a variety of imaging techniques and analyses. They then entered this data into a supercomputer to produce this image. Related to image 3477.
Juan R. Perilla and the Theoretical and Computational Biophysics Group, University of Illinois at Urbana-Champaign
View Media
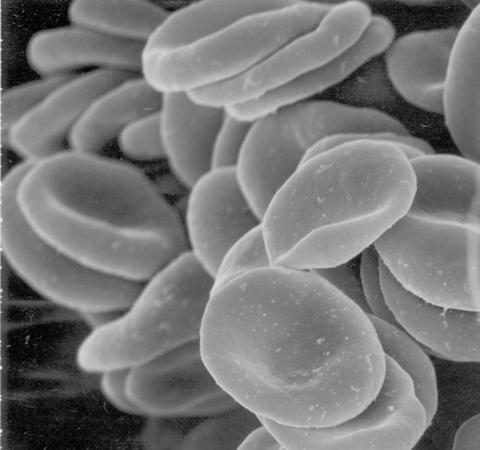
1101: Red blood cells
1101: Red blood cells
This image of human red blood cells was obtained with the help of a scanning electron microscope, an instrument that uses a finely focused electron beam to yield detailed images of the surface of a sample.
Tina Weatherby Carvalho, University of Hawaii at Manoa
View Media
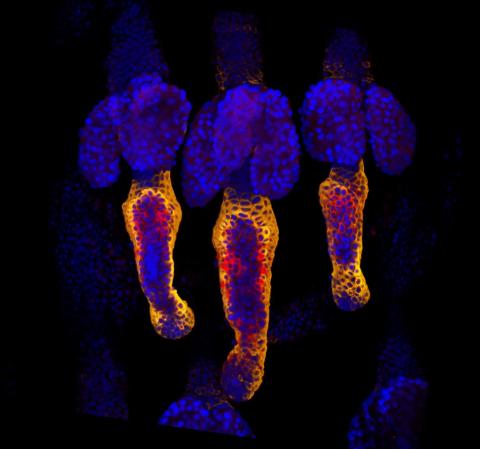
3499: Growing hair follicle stem cells
3499: Growing hair follicle stem cells
Wound healing requires the action of stem cells. In mice that lack the Sept2/ARTS gene, stem cells involved in wound healing live longer and wounds heal faster and more thoroughly than in normal mice. This confocal microscopy image from a mouse lacking the Sept2/ARTS gene shows a tail wound in the process of healing. Cell nuclei are in blue. Red and orange mark hair follicle stem cells (hair follicle stem cells activate to cause hair regrowth, which indicates healing). See more information in the article in Science.
Hermann Steller, Rockefeller University
View Media
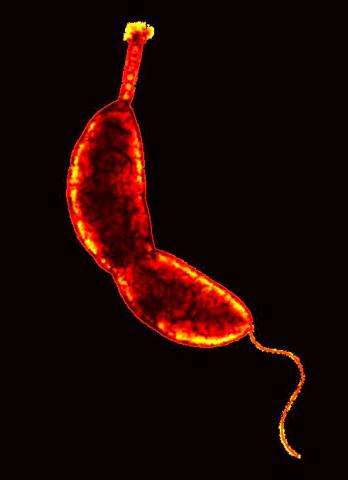
3262: Caulobacter
3262: Caulobacter
A study using Caulobacter crescentus showed that some bacteria use just-in-time processing, much like that used in industrial delivery, to make the glue that allows them to attach to surfaces, an important step in the infection process for many disease-causing bacteria. In the image shown, this freshwater bacterium has a holdfast at the top and a propelling flagellum at the end. From an Indiana University news release.
Yves Brun, Indiana University
View Media
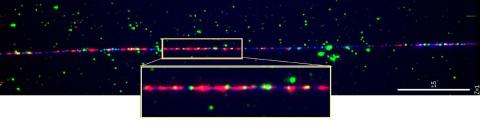
2475: Chromosome fiber 01
2475: Chromosome fiber 01
This microscopic image shows a chromatin fiber--a DNA molecule bound to naturally occurring proteins.
Marc Green and Susan Forsburg, University of Southern California
View Media
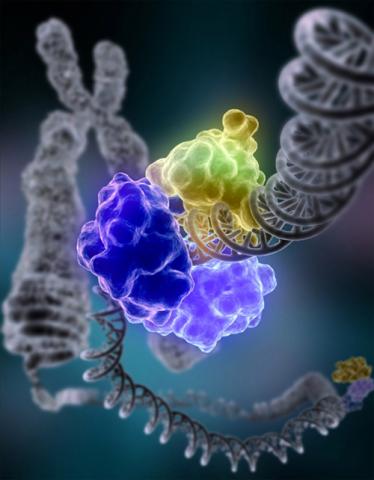
2330: Repairing DNA
2330: Repairing DNA
Like a watch wrapped around a wrist, a special enzyme encircles the double helix to repair a broken strand of DNA. Without molecules that can mend such breaks, cells can malfunction, die, or become cancerous. Related to image 3493.
Tom Ellenberger, Washington University School of Medicine
View Media
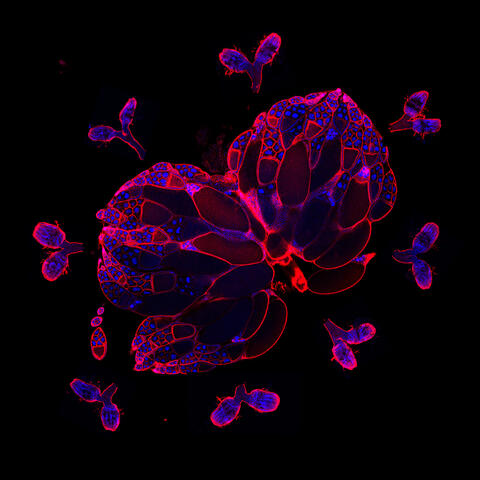
6806: Wild-type and mutant fruit fly ovaries
6806: Wild-type and mutant fruit fly ovaries
The two large, central, round shapes are ovaries from a typical fruit fly (Drosophila melanogaster). The small butterfly-like structures surrounding them are fruit fly ovaries where researchers suppressed the expression of a gene that controls microtubule polymerization and is necessary for normal development. This image was captured using a confocal laser scanning microscope.
Related to image 6807.
Related to image 6807.
Vladimir I. Gelfand, Feinberg School of Medicine, Northwestern University.
View Media
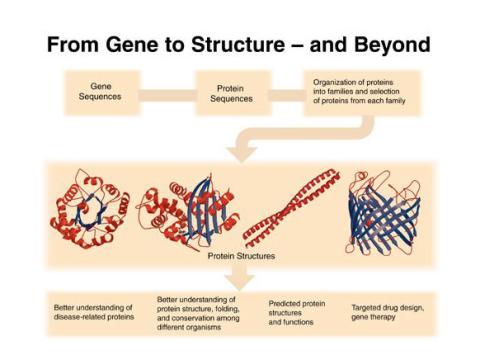
2363: PSI: from genes to structures
2363: PSI: from genes to structures
The goal of the Protein Structure Initiative (PSI) is to determine the three-dimensional shapes of a wide range of proteins by solving the structures of representative members of each protein family found in nature. The collection of structures should serve as a valuable resource for biomedical research scientists.
National Institute of General Medical Sciences
View Media

6609: 3D reconstruction of the Golgi apparatus in a pancreas cell
6609: 3D reconstruction of the Golgi apparatus in a pancreas cell
Researchers used cryo-electron tomography (cryo-ET) to capture images of a rat pancreas cell that were then compiled and color-coded to produce a 3D reconstruction. Visible features include the folded sacs of the Golgi apparatus (copper), transport vesicles (medium-sized dark-blue circles), microtubules (neon-green rods), a mitochondria membrane (pink), ribosomes (small pale-yellow circles), endoplasmic reticulum (aqua), and lysosomes (large yellowish-green circles). See 6606 for a still image from the video.
Xianjun Zhang, University of Southern California.
View Media
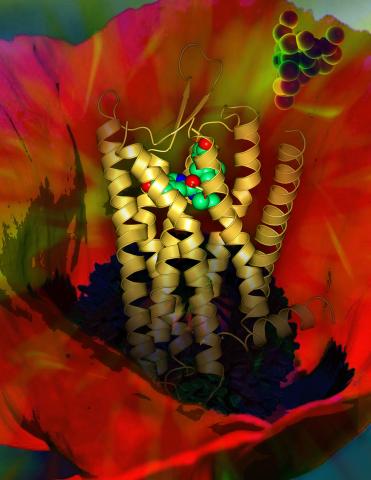
3314: Human opioid receptor structure superimposed on poppy
3314: Human opioid receptor structure superimposed on poppy
Opioid receptors on the surfaces of brain cells are involved in pleasure, pain, addiction, depression, psychosis, and other conditions. The receptors bind to both innate opioids and drugs ranging from hospital anesthetics to opium. Researchers at The Scripps Research Institute, supported by the NIGMS Protein Structure Initiative, determined the first three-dimensional structure of a human opioid receptor, a kappa-opioid receptor. In this illustration, the submicroscopic receptor structure is shown while bound to an agonist (or activator). The structure is superimposed on a poppy flower, the source of opium.
Raymond Stevens, The Scripps Research Institute
View Media
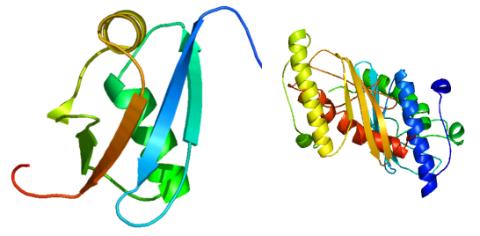
2388: Ubiquitin-fold modifier 1 from C. elegans
2388: Ubiquitin-fold modifier 1 from C. elegans
Solution NMR structure of protein target WR41 (left) from C. elegans. Noting the unanticipated structural similarity to the ubiquitin protein (Ub) found in all eukaryotic cells, researchers discovered that WR41 is a Ub-like modifier, ubiquitin-fold modifier 1 (Ufm1), on a newly uncovered ubiquitin-like pathway. Subsequently, the PSI group also determined the three-dimensional structure of protein target HR41 (right) from humans, the E2 ligase for Ufm1, using both NMR and X-ray crystallography.
Northeast Structural Genomics Consortium
View Media

2406: Hen egg lysozyme (2)
2406: Hen egg lysozyme (2)
A crystal of hen egg lysozyme protein created for X-ray crystallography, which can reveal detailed, three-dimensional protein structures.
Alex McPherson, University of California, Irvine
View Media
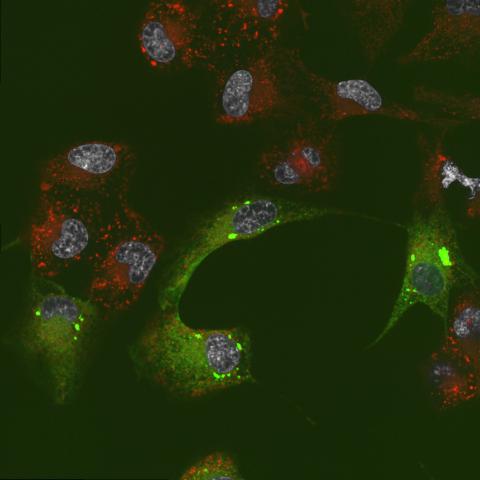
6774: Endoplasmic reticulum abnormalities 2
6774: Endoplasmic reticulum abnormalities 2
Human cells with the gene that codes for the protein FIT2 deleted. After an experimental intervention, they are expressing a nonfunctional version of FIT2, shown in green. The lack of functional FIT2 affected the structure of the endoplasmic reticulum (ER), and the nonfunctional protein clustered in ER membrane aggregates, seen as large bright-green spots. Lipid droplets are shown in red, and the nucleus is visible in gray. This image was captured using a confocal microscope. Related to image 6773.
Michel Becuwe, Harvard University.
View Media
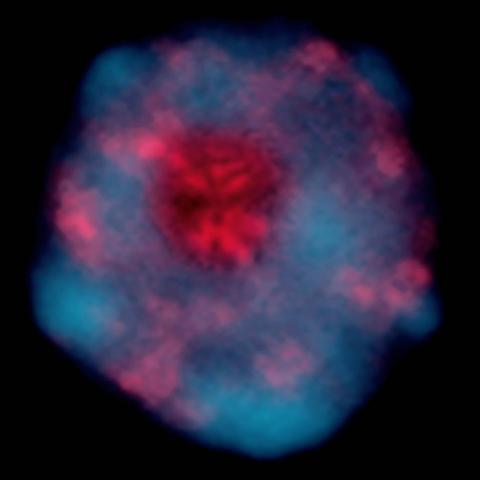
2318: Gene silencing
2318: Gene silencing
Pretty in pink, the enzyme histone deacetylase (HDA6) stands out against a background of blue-tinted DNA in the nucleus of an Arabidopsis plant cell. Here, HDA6 concentrates in the nucleolus (top center), where ribosomal RNA genes reside. The enzyme silences the ribosomal RNA genes from one parent while those from the other parent remain active. This chromosome-specific silencing of ribosomal RNA genes is an unusual phenomenon observed in hybrid plants.
Olga Pontes and Craig Pikaard, Washington University
View Media
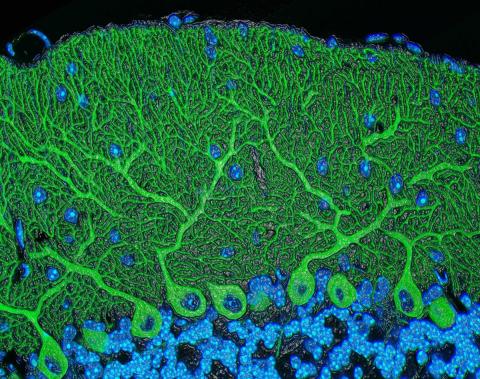
5795: Mouse cerebellum
5795: Mouse cerebellum
The cerebellum is the brain's locomotion control center. Found at the base of your brain, the cerebellum is a single layer of tissue with deep folds like an accordion. People with damage to this region of the brain often have difficulty with balance, coordination and fine motor skills.
This image of a mouse cerebellum is part of a collection of such images in different colors and at different levels of magnification from the National Center for Microscopy and Imaging Research (NCMIR). Related to image 5800.
This image of a mouse cerebellum is part of a collection of such images in different colors and at different levels of magnification from the National Center for Microscopy and Imaging Research (NCMIR). Related to image 5800.
National Center for Microscopy and Imaging Research (NCMIR)
View Media

5753: Clathrin-mediated endocytosis
5753: Clathrin-mediated endocytosis
Endocytosis is the process by which cells are able to take up membrane and extracellular materials through the formation of a small intracellular bubble, called a vesicle. This process, called membrane budding, is generally by a coating of proteins. This protein coat helps both to deform the membrane and to concentrate specific proteins inside the newly forming vesicle. Clathrin is a coat protein that functions in receptor-mediated endocytosis events at the plasma membrane. This animation shows the process of clathrin-mediated endocytosis. An iron-transport protein called transferrin (blue) is bound to its receptor (purple) on the exterior cell membrane. Inside the cell, a clathrin cage (shown in white/beige) assembles through interactions with membrane-bound adaptor proteins (green), causing the cell membrane to begin bending. The adaptor proteins also bind to receptors for transferrin, capturing them in the growing vesicle. Molecules of a protein called dynamin (purple) are then recruited to the neck of the vesicle and are involved in separating the membranes of the cell and the vesicle. Soon after the vesicle has budded off the membrane, the clathrin cage is disassembled. This disassembly is mediated by another protein called HSC70 (yellow), and its cofactor protein auxilin (orange).
Janet Iwasa, University of Utah
View Media
2439: Hydra 03
2439: Hydra 03
Hydra magnipapillata is an invertebrate animal used as a model organism to study developmental questions, for example the formation of the body axis.
Hiroshi Shimizu, National Institute of Genetics in Mishima, Japan
View Media
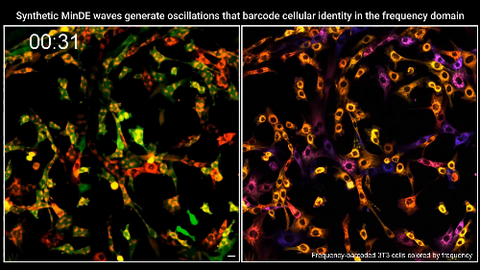
7022: Single-cell “radios” video
7022: Single-cell “radios” video
Individual cells are color-coded based on their identity and signaling activity using a protein circuit technology developed by the Coyle Lab. Just as a radio allows you to listen to an individual frequency, this technology allows researchers to tune into the specific “radio station” of each cell through genetically encoded proteins from a bacterial system called MinDE. The proteins generate an oscillating fluorescent signal that transmits information about cell shape, state, and identity that can be decoded using digital signal processing tools originally designed for telecommunications. The approach allows researchers to look at the dynamics of a single cell in the presence of many other cells.
Related to image 7021.
Related to image 7021.
Scott Coyle, University of Wisconsin-Madison.
View Media
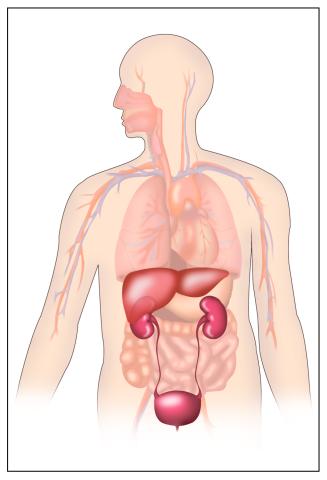
2496: Body toxins
2496: Body toxins
Body organs such as the liver and kidneys process chemicals and toxins. These "target" organs are susceptible to damage caused by these substances. See image 2497 for a labeled version of this illustration.
Crabtree + Company
View Media
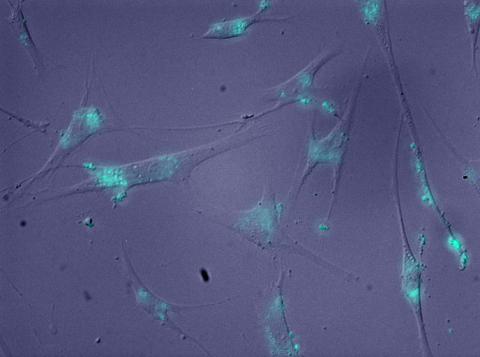
2600: Molecules blocking Huntington's protein production
2600: Molecules blocking Huntington's protein production
The molecules that glow blue in these cultured cells prevent the expression of the mutant proteins that cause Huntington's disease. Biochemist David Corey and others at UT Southwestern Medical Center designed the molecules to specifically target the genetic repeats that code for harmful proteins in people with Huntington's disese. People with Huntington's disease and similar neurodegenerative disorders often have extra copies of a gene segment. Moving from cell cultures to animals will help researchers further explore the potential of their specially crafted molecule to treat brain disorders. In addition to NIGMS, NIH's National Institute of Neurological Disorders and Stroke and National Institute of Biomedical Imaging and Bioengineering also funded this work.
Jiaxin Hu, David W. Dodd and Robert H. E. Hudson, UT Southwestern Medical Center
View Media
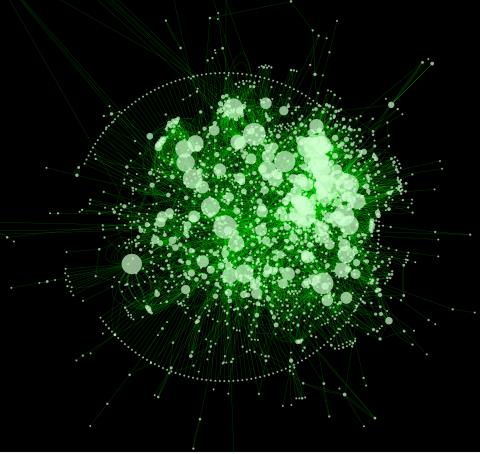
2737: Cytoscape network diagram 1
2737: Cytoscape network diagram 1
Molecular biologists are increasingly relying on bioinformatics software to visualize molecular interaction networks and to integrate these networks with data such as gene expression profiles. Related to 2749.
Keiichiro Ono, Trey Ideker lab, University of California, San Diego
View Media
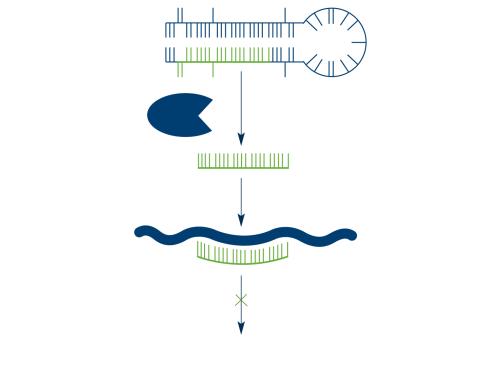
2556: Dicer generates microRNAs
2556: Dicer generates microRNAs
The enzyme Dicer generates microRNAs by chopping larger RNA molecules into tiny Velcro®-like pieces. MicroRNAs stick to mRNA molecules and prevent the mRNAs from being made into proteins. See image 2557 for a labeled version of this illustration. Featured in The New Genetics.
Crabtree + Company
View Media
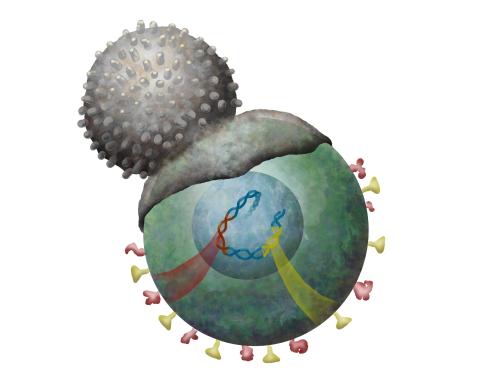
2489: Immune cell attacks cell infected with a retrovirus
2489: Immune cell attacks cell infected with a retrovirus
T cells engulf and digest cells displaying markers (or antigens) for retroviruses, such as HIV.
Kristy Whitehouse, science illustrator
View Media
1084: Natcher Building 04
1084: Natcher Building 04
NIGMS staff are located in the Natcher Building on the NIH campus.
Alisa Machalek, National Institute of General Medical Sciences
View Media
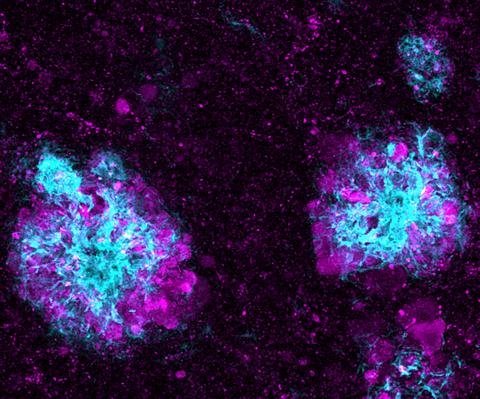
5771: Lysosome clusters around amyloid plaques
5771: Lysosome clusters around amyloid plaques
It's probably most people's least favorite activity, but we still need to do it--take out our trash. Otherwise our homes will get cluttered and smelly, and eventually, we'll get sick. The same is true for our cells: garbage disposal is an ongoing and essential activity, and our cells have a dedicated waste-management system that helps keep them clean and neat. One major waste-removal agent in the cell is the lysosome. Lysosomes are small structures, called organelles, and help the body to dispose of proteins and other molecules that have become damaged or worn out.
This image shows a massive accumulation of lysosomes (visualized with LAMP1 immunofluorescence, in purple) within nerve cells that surround amyloid plaques (visualized with beta-amyloid immunofluorescence, in light blue) in a mouse model of Alzheimer's disease. Scientists have linked accumulation of lysosomes around amyloid plaques to impaired waste disposal in nerve cells, ultimately resulting in cell death.
This image shows a massive accumulation of lysosomes (visualized with LAMP1 immunofluorescence, in purple) within nerve cells that surround amyloid plaques (visualized with beta-amyloid immunofluorescence, in light blue) in a mouse model of Alzheimer's disease. Scientists have linked accumulation of lysosomes around amyloid plaques to impaired waste disposal in nerve cells, ultimately resulting in cell death.
Swetha Gowrishankar and Shawn Ferguson, Yale School of Medicine
View Media
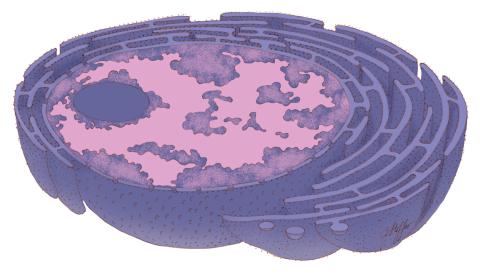
1290: Nucleus and rough ER
1290: Nucleus and rough ER
The nucleus contains the DNA of eukaryotic cells. The double membrane that bounds the nucleus flows into the rough endoplasmic reticulum, an organelle studded with ribosomes that manufacture membrane-bound proteins for the rest of the cell.
Judith Stoffer
View Media
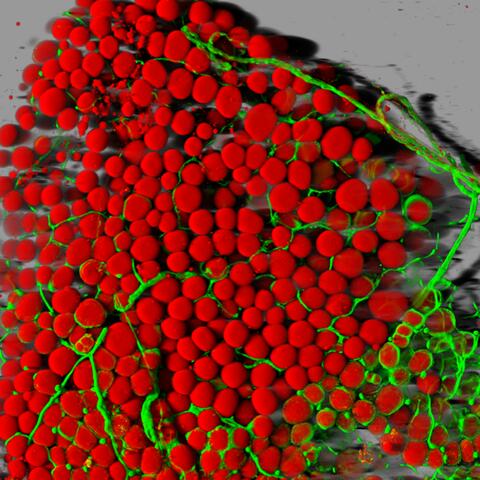
3600: Fat cells (red) and blood vessels (green)
3600: Fat cells (red) and blood vessels (green)
A mouse's fat cells (red) are shown surrounded by a network of blood vessels (green). Fat cells store and release energy, protect organs and nerve tissues, insulate us from the cold, and help us absorb important vitamins.
This image was part of the Life: Magnified exhibit that ran from June 3, 2014, to January 21, 2015, at Dulles International Airport.
This image was part of the Life: Magnified exhibit that ran from June 3, 2014, to January 21, 2015, at Dulles International Airport.
Daniela Malide, National Heart, Lung, and Blood Institute, National Institutes of Health
View Media
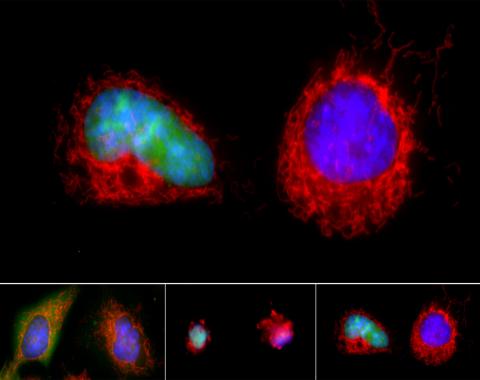
3486: Apoptosis reversed
3486: Apoptosis reversed
Two healthy cells (bottom, left) enter into apoptosis (bottom, center) but spring back to life after a fatal toxin is removed (bottom, right; top).
Hogan Tang of the Denise Montell Lab, Johns Hopkins University School of Medicine
View Media
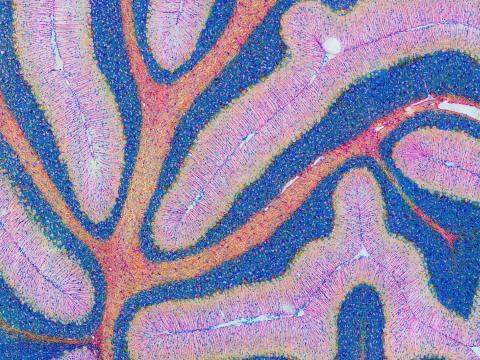
5800: Mouse cerebellum in pink and blue
5800: Mouse cerebellum in pink and blue
The cerebellum is the brain's locomotion control center. Found at the base of your brain, the cerebellum is a single layer of tissue with deep folds like an accordion. People with damage to this region of the brain often have difficulty with balance, coordination and fine motor skills.
This image of a mouse cerebellum is part of a collection of such images in different colors and at different levels of magnification from the National Center for Microscopy and Imaging Research (NCMIR). Related to image 5795.
This image of a mouse cerebellum is part of a collection of such images in different colors and at different levels of magnification from the National Center for Microscopy and Imaging Research (NCMIR). Related to image 5795.
National Center for Microscopy and Imaging Research (NCMIR)
View Media
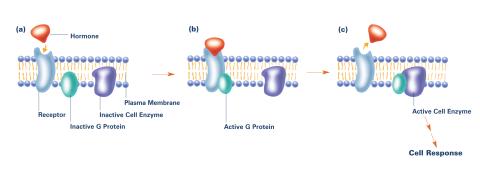
2538: G switch (with labels and stages)
2538: G switch (with labels and stages)
The G switch allows our bodies to respond rapidly to hormones. G proteins act like relay batons to pass messages from circulating hormones into cells. A hormone (red) encounters a receptor (blue) in the membrane of a cell. Next, a G protein (green) becomes activated and makes contact with the receptor to which the hormone is attached. Finally, the G protein passes the hormone's message to the cell by switching on a cell enzyme (purple) that triggers a response. See image 2536 and 2537 for other versions of this image. Featured in Medicines By Design.
Crabtree + Company
View Media
2715: Glow-in-the-dark salamanders
2715: Glow-in-the-dark salamanders
These six-month-old axolotls, a kind of salamander, glow green and blue under ultraviolet light. That's because they were genetically modified to make harmless green fluorescent protein, or GFP. Like X-ray vision, GFP lets you see inside the axolotls as they hang out in their aquarium. GFP not only can reveal internal structures in living organisms, but it also can light up specific cells and even proteins within a cell. That allows scientists to identify and track things like cancer cells.
View Media
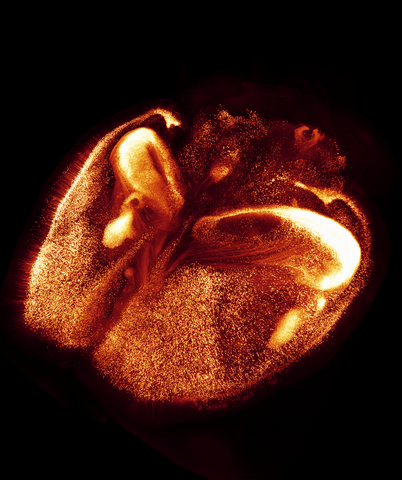
6930: Mouse brain 2
6930: Mouse brain 2
A mouse brain that was genetically modified so that subpopulations of its neurons glow. Researchers often study mice because they share many genes with people and can shed light on biological processes, development, and diseases in humans.
This image was captured using a light sheet microscope.
Related to image 6929 and video 6931.
This image was captured using a light sheet microscope.
Related to image 6929 and video 6931.
Prayag Murawala, MDI Biological Laboratory and Hannover Medical School.
View Media

2714: Stretch detectors
2714: Stretch detectors
Muscles stretch and contract when we walk, and skin splits open and knits back together when we get a paper cut. To study these contractile forces, researchers built a three-dimensional scaffold that mimics tissue in an organism. Researchers poured a mixture of cells and elastic collagen over microscopic posts in a dish. Then they studied how the cells pulled and released the posts as they formed a web of tissue. To measure forces between posts, the researchers developed a computer model. Their findings--which show that contractile forces vary throughout the tissue--could have a wide range of medical applications.
Christopher Chen, University of Pennsylvania
View Media
1082: Natcher Building 02
1082: Natcher Building 02
NIGMS staff are located in the Natcher Building on the NIH campus.
Alisa Machalek, National Institute of General Medical Sciences
View Media
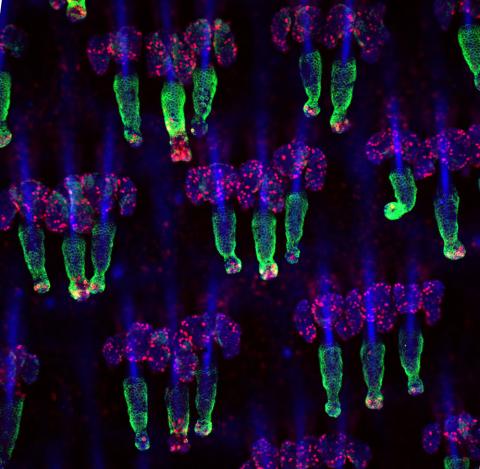
3498: Wound healing in process
3498: Wound healing in process
Wound healing requires the action of stem cells. In mice that lack the Sept2/ARTS gene, stem cells involved in wound healing live longer and wounds heal faster and more thoroughly than in normal mice. This confocal microscopy image from a mouse lacking the Sept2/ARTS gene shows a tail wound in the process of healing. See more information in the article in Science.
Related to images 3497 and 3500.
Related to images 3497 and 3500.
Hermann Steller, Rockefeller University
View Media
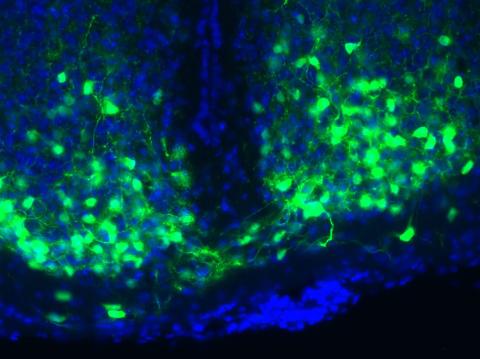
3547: Master clock of the mouse brain
3547: Master clock of the mouse brain
An image of the area of the mouse brain that serves as the 'master clock,' which houses the brain's time-keeping neurons. The nuclei of the clock cells are shown in blue. A small molecule called VIP, shown in green, enables neurons in the central clock in the mammalian brain to synchronize.
Erik Herzog, Washington University in St. Louis
View Media