Switch to List View
Image and Video Gallery
This is a searchable collection of scientific photos, illustrations, and videos. The images and videos in this gallery are licensed under Creative Commons Attribution Non-Commercial ShareAlike 3.0. This license lets you remix, tweak, and build upon this work non-commercially, as long as you credit and license your new creations under identical terms.
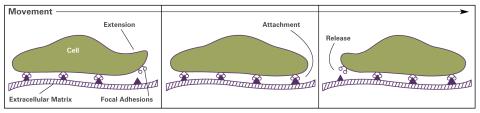
2503: Focal adhesions (with labels)
2503: Focal adhesions (with labels)
Cells walk along body surfaces via tiny "feet," called focal adhesions, that connect with the extracellular matrix. See image 2502 for an unlabeled version of this illustration.
Crabtree + Company
View Media

3735: Scanning electron microscopy of collagen fibers
3735: Scanning electron microscopy of collagen fibers
This image shows collagen, a fibrous protein that's the main component of the extracellular matrix (ECM). Collagen is a strong, ropelike molecule that forms stretch-resistant fibers. The most abundant protein in our bodies, collagen accounts for about a quarter of our total protein mass. Among its many functions is giving strength to our tendons, ligaments and bones and providing scaffolding for skin wounds to heal. There are about 20 different types of collagen in our bodies, each adapted to the needs of specific tissues.
Tom Deerinck, National Center for Microscopy and Imaging Research (NCMIR)
View Media
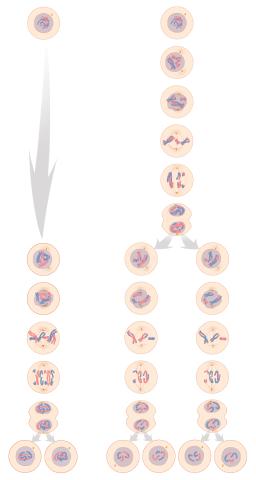
1333: Mitosis and meiosis compared
1333: Mitosis and meiosis compared
Meiosis is used to make sperm and egg cells. During meiosis, a cell's chromosomes are copied once, but the cell divides twice. During mitosis, the chromosomes are copied once, and the cell divides once. For simplicity, cells are illustrated with only three pairs of chromosomes. See image 6788 for a labeled version of this illustration.
Judith Stoffer
View Media

2335: Virtual snow world
2335: Virtual snow world
Glide across an icy canyon, where you see smiling snowmen and waddling penguins. Toss a snowball, hear it smash against an igloo, and then watch it explode in bright colors. Psychologists David Patterson and Hunter Hoffman of the University of Washington in Seattle developed this virtual "Snow World" to test whether immersing someone in a pretend reality could ease pain during burn treatment and other medical procedures. They found that people fully engaged in the virtual reality experience reported 60 percent less pain. The technology offers a promising way to manage pain.
David Patterson and Hunter Hoffmann, University of Washington
View Media
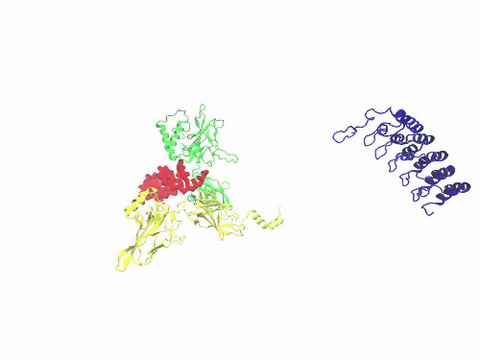
3729: A molecular switch strips transcription factor from DNA
3729: A molecular switch strips transcription factor from DNA
In this video, Rice University scientists used molecular modeling with a mathematical algorithm called AWSEM (for associative memory, water-mediated, structure and energy model) and structural data to analyze how a transcription factor called nuclear factor kappa B (NFkB) is removed from DNA to stop gene activation. AWSEM uses the interacting energies of their components to predict how proteins fold. At the start, the NFkB dimer (green and yellow, in the center) grips DNA (red, to the left), which activates the transcription of genes. IkB (blue, to the right), an inhibitor protein, stops transcription when it binds to NFkB and forces the dimer to twist and release its hold on DNA. The yellow domain at the bottom of IkB is the PEST domain, which binds first to NFkB. For more details about this mechanism called molecular stripping, see here.
Davit Potoyan and Peter Wolynes
View Media
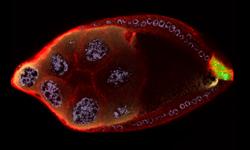
2309: Cellular polarity
2309: Cellular polarity
As an egg cell develops, a process called polarization controls what parts ultimately become the embryo's head and tail. This picture shows an egg of the fruit fly Drosophila. Red and green mark two types of signaling proteins involved in polarization. Disrupting these signals can scramble the body plan of the embryo, leading to severe developmental disorders.
Wu-Min Deng, Florida State University
View Media
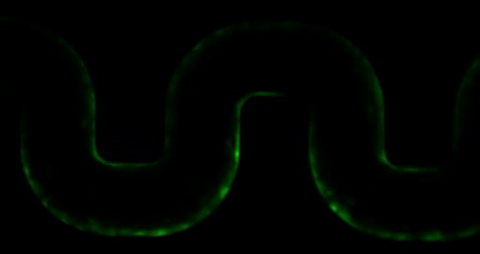
3446: Biofilm blocking fluid flow
3446: Biofilm blocking fluid flow
This time-lapse movie shows that bacterial communities called biofilms can create blockages that prevent fluid flow in devices such as stents and catheters over a period of about 56 hours. This video was featured in a news release from Princeton University.
Bonnie Bassler, Princeton University
View Media
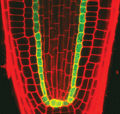
2329: Planting roots
2329: Planting roots
At the root tips of the mustard plant Arabidopsis thaliana (red), two proteins work together to control the uptake of water and nutrients. When the cell division-promoting protein called Short-root moves from the center of the tip outward, it triggers the production of another protein (green) that confines Short-root to the nutrient-filtering endodermis. The mechanism sheds light on how genes and proteins interact in a model organism and also could inform the engineering of plants.
Philip Benfey, Duke University
View Media
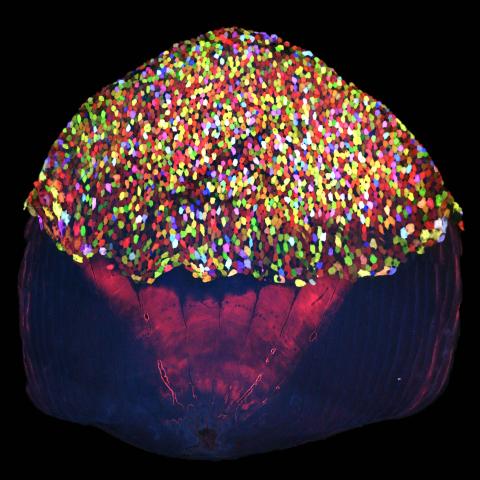
3783: A multicolored fish scale 2
3783: A multicolored fish scale 2
Each of the tiny colored specs in this image is a cell on the surface of a fish scale. To better understand how wounds heal, scientists have inserted genes that make cells brightly glow in different colors into the skin cells of zebrafish, a fish often used in laboratory research. The colors enable the researchers to track each individual cell, for example, as it moves to the location of a cut or scrape over the course of several days. These technicolor fish endowed with glowing skin cells dubbed "skinbow" provide important insight into how tissues recover and regenerate after an injury.
For more information on skinbow fish, see the Biomedical Beat blog post Visualizing Skin Regeneration in Real Time and a press release from Duke University highlighting this research. Related to image 3782.
For more information on skinbow fish, see the Biomedical Beat blog post Visualizing Skin Regeneration in Real Time and a press release from Duke University highlighting this research. Related to image 3782.
Chen-Hui Chen and Kenneth Poss, Duke University
View Media
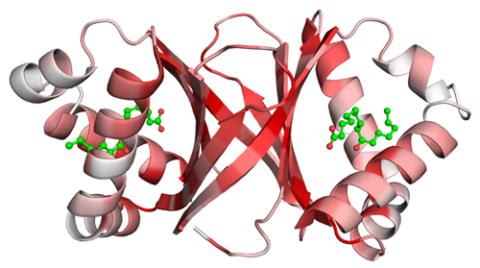
2340: Dimeric ferredoxin-like protein from an unidentified marine microbe
2340: Dimeric ferredoxin-like protein from an unidentified marine microbe
This is the first structure of a protein derived from the metagenomic sequences collected during the Sorcerer II Global Ocean Sampling project. The crystal structure shows a barrel protein with a ferredoxin-like fold and a long chain fatty acid in a deep cleft (shaded red). Featured as one of the August 2007 Protein Structure Initiative Structures of the Month.
Joint Center for Structural Genomics
View Media

6804: Staphylococcus aureus in the porous coating of a femoral hip stem
6804: Staphylococcus aureus in the porous coating of a femoral hip stem
Staphylococcus aureus bacteria (blue) on the porous coating of a femoral hip stem used in hip replacement surgery. The relatively rough surface of an implant is a favorable environment for bacteria to attach and grow. This can lead to the development of biofilms, which can cause infections. The researchers who took this image are working to understand where biofilms are likely to develop. This knowledge could support the prevention and treatment of infections. A scanning electron microscope was used to capture this image.
More information on the research that produced this image can be found in the Antibiotics paper "Free-floating aggregate and single-cell-initiated biofilms of Staphylococcus aureus" by Gupta et al.
Related to image 6803 and video 6805.
More information on the research that produced this image can be found in the Antibiotics paper "Free-floating aggregate and single-cell-initiated biofilms of Staphylococcus aureus" by Gupta et al.
Related to image 6803 and video 6805.
Paul Stoodley, The Ohio State University.
View Media
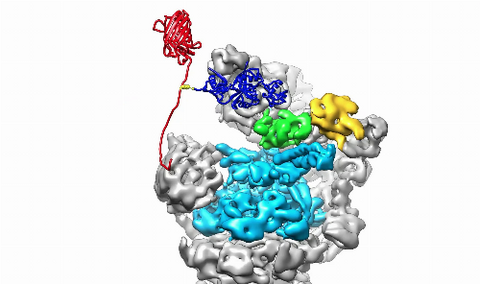
3764: Movie of the 19S proteasome subunit processing a protein substrate
3764: Movie of the 19S proteasome subunit processing a protein substrate
The proteasome is a critical multiprotein complex in the cell that breaks down and recycles proteins that have become damaged or are no longer needed. This movie shows how a protein substrate (red) is bound through its ubiquitin chain (blue) to one of the ubiquitin receptors of the proteasome (Rpn10, yellow). The substrate's flexible engagement region then gets engaged by the AAA+ motor of the proteasome (cyan), which initiates mechanical pulling, unfolding and movement of the protein into the proteasome's interior for cleavage into shorter protein pieces called peptides. During movement of the substrate, its ubiquitin modification gets cleaved off by the deubiquitinase Rpn11 (green), which sits directly above the entrance to the AAA+ motor pore and acts as a gatekeeper to ensure efficient ubiquitin removal, a prerequisite for fast protein breakdown by the 26S proteasome. Related to image 3763.
Andreas Martin, HHMI
View Media
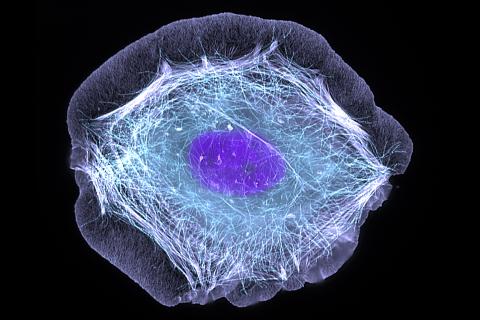
3599: Skin cell (keratinocyte)
3599: Skin cell (keratinocyte)
This normal human skin cell was treated with a growth factor that triggered the formation of specialized protein structures that enable the cell to move. We depend on cell movement for such basic functions as wound healing and launching an immune response.
This image was part of the Life: Magnified exhibit that ran from June 3, 2014, to January 21, 2015, at Dulles International Airport.
This image was part of the Life: Magnified exhibit that ran from June 3, 2014, to January 21, 2015, at Dulles International Airport.
Torsten Wittmann, University of California, San Francisco
View Media
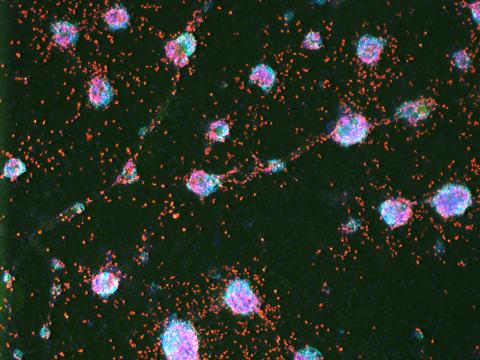
3277: Human ES cells turn into insulin-producing cells
3277: Human ES cells turn into insulin-producing cells
Human embryonic stem cells were differentiated into cells like those found in the pancreas (blue), which give rise to insulin-producing cells (red). When implanted in mice, the stem cell-derived pancreatic cells can replace the insulin that isn't produced in type 1 diabetes. Image and caption information courtesy of the California Institute for Regenerative Medicine.
Eugene Brandon, ViaCyte, via CIRM
View Media
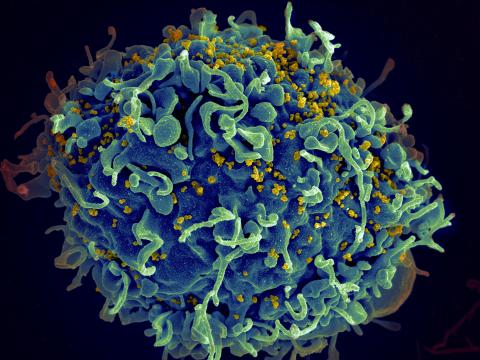
3638: HIV, the AIDS virus, infecting a human cell
3638: HIV, the AIDS virus, infecting a human cell
This human T cell (blue) is under attack by HIV (yellow), the virus that causes AIDS. The virus specifically targets T cells, which play a critical role in the body's immune response against invaders like bacteria and viruses.
This image was part of the Life: Magnified exhibit that ran from June 3, 2014, to January 21, 2015, at Dulles International Airport.
This image was part of the Life: Magnified exhibit that ran from June 3, 2014, to January 21, 2015, at Dulles International Airport.
Seth Pincus, Elizabeth Fischer, and Austin Athman, National Institute of Allergy and Infectious Diseases, National Institutes of Health
View Media

5810: Tongue 1
5810: Tongue 1
Microscopy image of tongue. One in a series of two, see image 5811
National Center for Microscopy and Imaging Research (NCMIR)
View Media
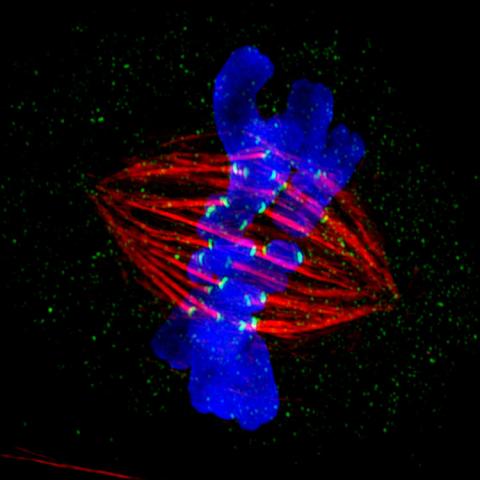
3445: Dividing cell in metaphase
3445: Dividing cell in metaphase
This image of a mammalian epithelial cell, captured in metaphase, was the winning image in the high- and super-resolution microscopy category of the 2012 GE Healthcare Life Sciences Cell Imaging Competition. The image shows microtubules (red), kinetochores (green) and DNA (blue). The DNA is fixed in the process of being moved along the microtubules that form the structure of the spindle.
The image was taken using the DeltaVision OMX imaging system, affectionately known as the "OMG" microscope, and was displayed on the NBC screen in New York's Times Square during the weekend of April 20-21, 2013. It was also part of the Life: Magnified exhibit that ran from June 3, 2014, to January 21, 2015, at Dulles International Airport.
The image was taken using the DeltaVision OMX imaging system, affectionately known as the "OMG" microscope, and was displayed on the NBC screen in New York's Times Square during the weekend of April 20-21, 2013. It was also part of the Life: Magnified exhibit that ran from June 3, 2014, to January 21, 2015, at Dulles International Airport.
Jane Stout in the laboratory of Claire Walczak, Indiana University, GE Healthcare 2012 Cell Imaging Competition
View Media
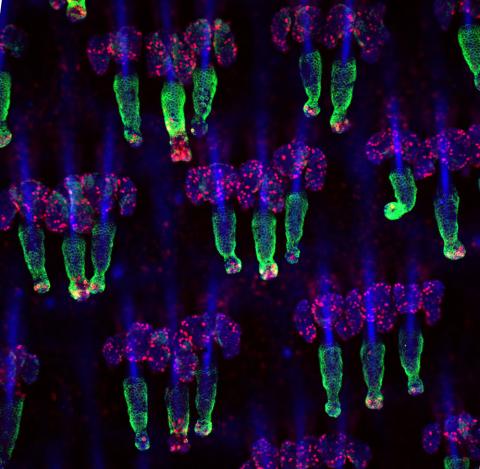
3498: Wound healing in process
3498: Wound healing in process
Wound healing requires the action of stem cells. In mice that lack the Sept2/ARTS gene, stem cells involved in wound healing live longer and wounds heal faster and more thoroughly than in normal mice. This confocal microscopy image from a mouse lacking the Sept2/ARTS gene shows a tail wound in the process of healing. See more information in the article in Science.
Related to images 3497 and 3500.
Related to images 3497 and 3500.
Hermann Steller, Rockefeller University
View Media
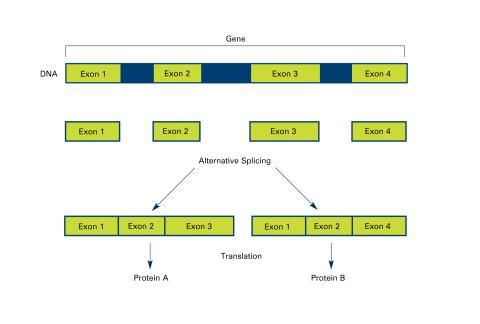
2553: Alternative splicing (with labels)
2553: Alternative splicing (with labels)
Arranging exons in different patterns, called alternative splicing, enables cells to make different proteins from a single gene. Featured in The New Genetics.
See image 2552 for an unlabeled version of this illustration.
See image 2552 for an unlabeled version of this illustration.
Crabtree + Company
View Media
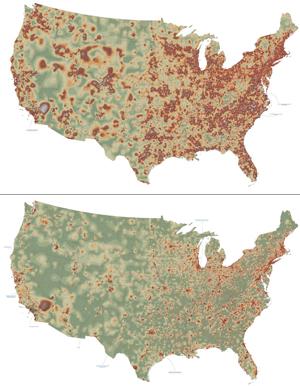
2320: Mapping disease spread
2320: Mapping disease spread
How far and fast an infectious disease spreads across a community depends on many factors, including transportation. These U.S. maps, developed as part of an international study to simulate and analyze disease spread, chart daily commuting patterns. They show where commuters live (top) and where they travel for work (bottom). Green represents the fewest number of people whereas orange, brown, and white depict the most. Such information enables researchers and policymakers to visualize how an outbreak in one area can spread quickly across a geographic region.
David Chrest, RTI International
View Media
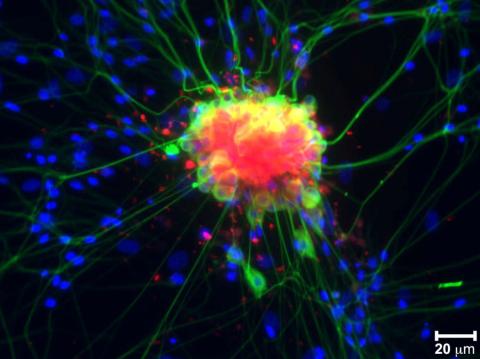
3251: Spinal nerve cells
3251: Spinal nerve cells
Neurons (green) and glial cells from isolated dorsal root ganglia express COX-2 (red) after exposure to an inflammatory stimulus (cell nuclei are blue). Lawrence Marnett and colleagues have demonstrated that certain drugs selectively block COX-2 metabolism of endocannabinoids -- naturally occurring analgesic molecules -- in stimulated dorsal root ganglia. Featured in the October 20, 2011 issue of Biomedical Beat.
Lawrence Marnett, Vanderbilt University
View Media
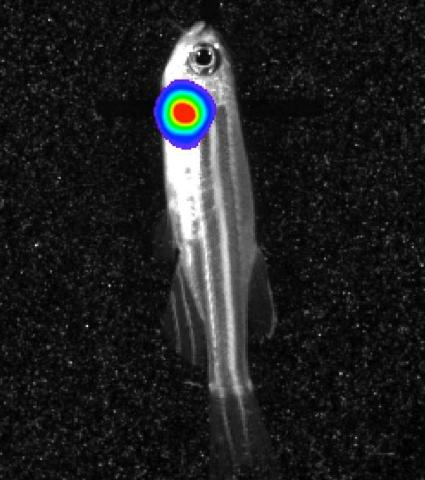
3558: Bioluminescent imaging in adult zebrafish - lateral view
3558: Bioluminescent imaging in adult zebrafish - lateral view
Luciferase-based imaging enables visualization and quantification of internal organs and transplanted cells in live adult zebrafish. In this image, a cardiac muscle-restricted promoter drives firefly luciferase expression (lateral view).
For imagery of both the lateral and overhead view go to 3556.
For imagery of the overhead view go to 3557.
For more information about the illumated area go to 3559.
For imagery of both the lateral and overhead view go to 3556.
For imagery of the overhead view go to 3557.
For more information about the illumated area go to 3559.
Kenneth Poss, Duke University
View Media
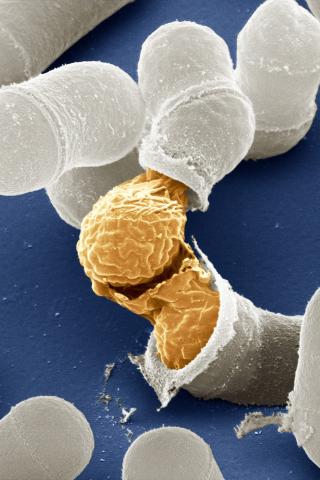
3614: Birth of a yeast cell
3614: Birth of a yeast cell
Yeast make bread, beer, and wine. And like us, yeast can reproduce sexually. A mother and father cell fuse and create one large cell that contains four offspring. When environmental conditions are favorable, the offspring are released, as shown here. Yeast are also a popular study subject for scientists. Research on yeast has yielded vast knowledge about basic cellular and molecular biology as well as about myriad human diseases, including colon cancer and various metabolic disorders.
This image was part of the Life: Magnified exhibit that ran from June 3, 2014, to January 21, 2015, at Dulles International Airport.
This image was part of the Life: Magnified exhibit that ran from June 3, 2014, to January 21, 2015, at Dulles International Airport.
Juergen Berger, Max Planck Institute for Developmental Biology, and Maria Langegger, Friedrich Miescher Laboratory of the Max Planck Society, Germany
View Media
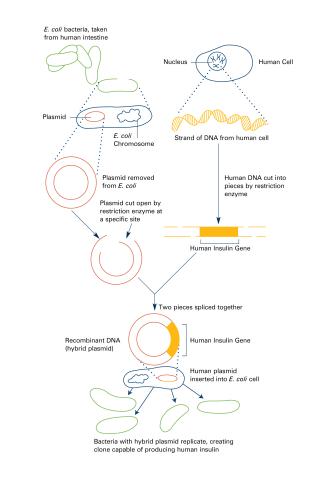
2565: Recombinant DNA (with labels)
2565: Recombinant DNA (with labels)
To splice a human gene (in this case, the one for insulin) into a plasmid, scientists take the plasmid out of an E. coli bacterium, cut the plasmid with a restriction enzyme, and splice in insulin-making human DNA. The resulting hybrid plasmid can be inserted into another E. coli bacterium, where it multiplies along with the bacterium. There, it can produce large quantities of insulin. See image 2564 for an unlabeled version of this illustration. Featured in The New Genetics.
Crabtree + Company
View Media
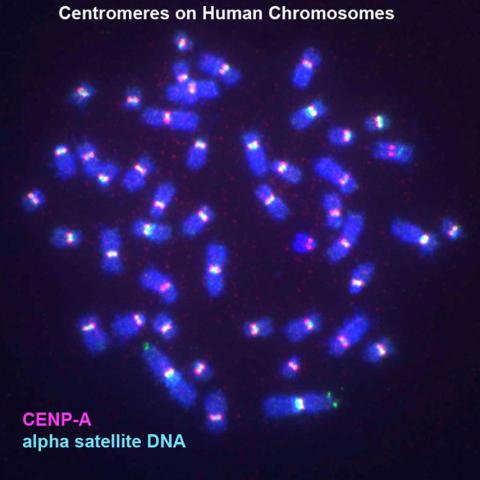
3255: Centromeres on human chromosomes
3255: Centromeres on human chromosomes
Human metaphase chromosomes are visible with fluorescence in vitro hybridization (FISH). Centromeric alpha satellite DNA (green) are found in the heterochromatin at each centromere. Immunofluorescence with CENP-A (red) shows the centromere-specific histone H3 variant that specifies the kinetochore.
Peter Warburton, Mount Sinai School of Medicine
View Media
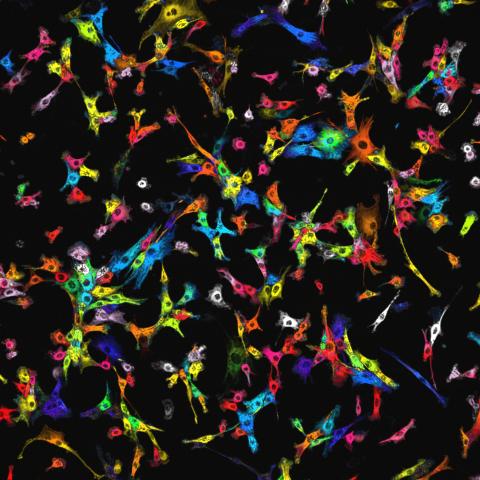
7021: Single-cell “radios” image
7021: Single-cell “radios” image
Individual cells are color-coded based on their identity and signaling activity using a protein circuit technology developed by the Coyle Lab. Just as a radio allows you to listen to an individual frequency, this technology allows researchers to tune into the specific “radio station” of each cell through genetically encoded proteins from a bacterial system called MinDE. The proteins generate an oscillating fluorescent signal that transmits information about cell shape, state, and identity that can be decoded using digital signal processing tools originally designed for telecommunications. The approach allows researchers to look at the dynamics of a single cell in the presence of many other cells.
Related to video 7022.
Related to video 7022.
Scott Coyle, University of Wisconsin-Madison.
View Media
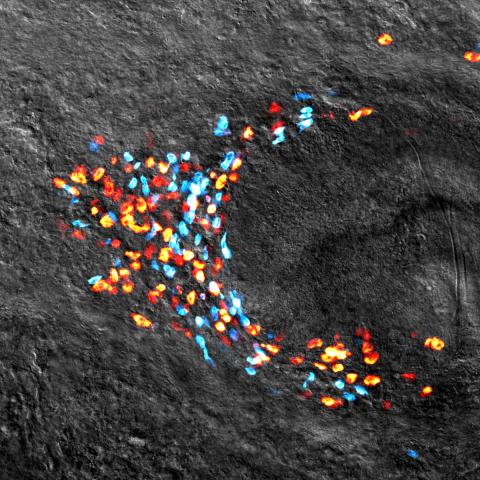
3306: Planarian stem cell colony
3306: Planarian stem cell colony
Planarians are freshwater flatworms that have powerful abilities to regenerate their bodies, which would seem to make them natural model organisms in which to study stem cells. But until recently, scientists had not been able to efficiently find the genes that regulate the planarian stem cell system. In this image, a single stem cell has given rise to a colony of stem cells in a planarian. Proliferating cells are red, and differentiating cells are blue. Quantitatively measuring the size and ratios of these two cell types provides a powerful framework for studying the roles of stem cell regulatory genes in planarians.
Peter Reddien, Whitehead Institute
View Media
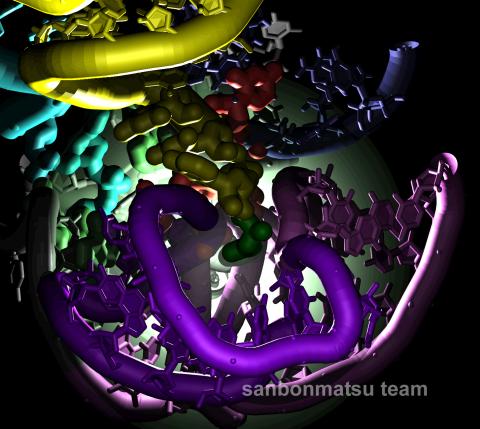
2336: Natural nanomachine in action
2336: Natural nanomachine in action
Using a supercomputer to simulate the movement of atoms in a ribosome, researchers looked into the core of this protein-making nanomachine and took snapshots. The picture shows an amino acid (green) being delivered by transfer RNA (yellow) into a corridor (purple) in the ribosome. In the corridor, a series of chemical reactions will string together amino acids to make a protein. The research project, which tracked the movement of more than 2.6 million atoms, was the largest computer simulation of a biological structure to date. The results shed light on the manufacturing of proteins and could aid the search for new antibiotics, which typically work by disabling the ribosomes of bacteria.
Kevin Sanbonmatsu, Los Alamos National Laboratory
View Media
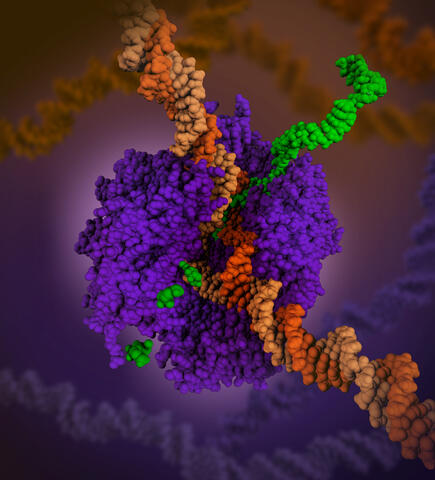
6993: RNA polymerase
6993: RNA polymerase
RNA polymerase (purple) is a complex enzyme at the heart of transcription. During this process, the enzyme unwinds the DNA double helix and uses one strand (darker orange) as a template to create the single-stranded messenger RNA (green), later used by ribosomes for protein synthesis.
From the RNA polymerase II elongation complex of Saccharomyces cerevisiae (PDB entry 1I6H) as seen in PDB-101's What is a Protein? video.
From the RNA polymerase II elongation complex of Saccharomyces cerevisiae (PDB entry 1I6H) as seen in PDB-101's What is a Protein? video.
Amy Wu and Christine Zardecki, RCSB Protein Data Bank.
View Media
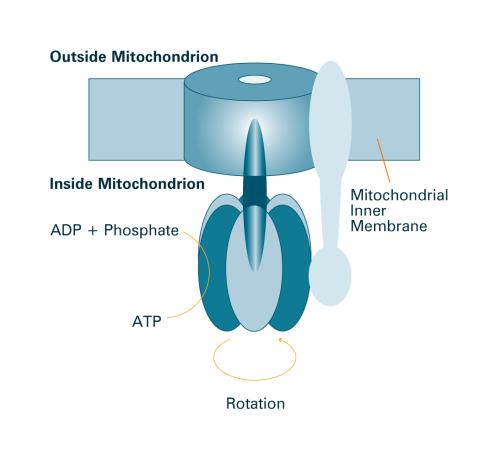
2518: ATP synthase (with labels)
2518: ATP synthase (with labels)
The world's smallest motor, ATP synthase, generates energy for the cell. See image 2517 for an unlabeled version of this illustration. Featured in The Chemistry of Health.
Crabtree + Company
View Media
2722: Cryogenic storage tanks at the Coriell Institute for Medical Research
2722: Cryogenic storage tanks at the Coriell Institute for Medical Research
Established in 1953, the Coriell Institute for Medical Research distributes cell lines and DNA samples to researchers around the world. Shown here are Coriell's cryogenic tanks filled with liquid nitrogen and millions of vials of frozen cells.
Courtney Sill, Coriell Institute for Medical Research
View Media
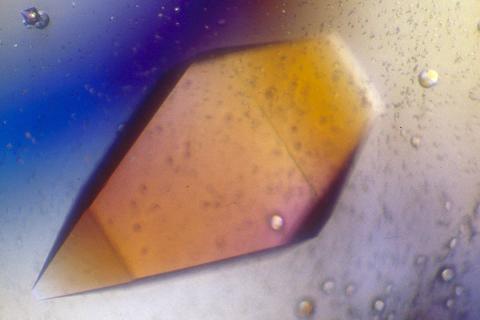
2400: Pig trypsin (1)
2400: Pig trypsin (1)
A crystal of porcine trypsin protein created for X-ray crystallography, which can reveal detailed, three-dimensional protein structures.
Alex McPherson, University of California, Irvine
View Media

2326: Nano-rainbow
2326: Nano-rainbow
These vials may look like they're filled with colored water, but they really contain nanocrystals reflecting different colors under ultraviolet light. The tiny crystals, made of semiconducting compounds, are called quantum dots. Depending on their size, the dots emit different colors that let scientists use them as a tool for detecting particular genes, proteins, and other biological molecules.
Shuming Nie, Emory University
View Media
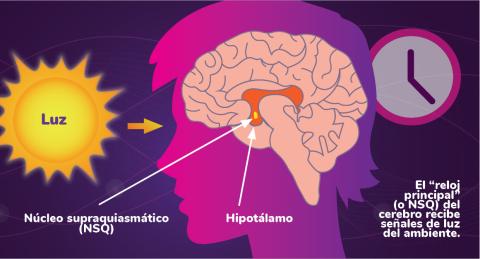
6614: Los ritmos circadianos y el núcleo supraquiasmático
6614: Los ritmos circadianos y el núcleo supraquiasmático
Los ritmos circadianos son cambios físicos, mentales y de comportamiento que siguen un ciclo de 24 horas. Los ritmos circadianos se ven influenciados por la luz y están regulados por el núcleo supraquiasmático del cerebro, a veces denominado el reloj principal.
Vea 6613 para la versión en inglés de esta infografía.
Vea 6613 para la versión en inglés de esta infografía.
NIGMS
View Media
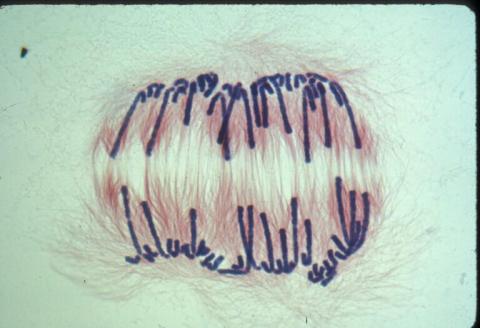
1010: Lily mitosis 10
1010: Lily mitosis 10
A light microscope image of a cell from the endosperm of an African globe lily (Scadoxus katherinae). This is one frame of a time-lapse sequence that shows cell division in action. The lily is considered a good organism for studying cell division because its chromosomes are much thicker and easier to see than human ones. Staining shows microtubules in red and chromosomes in blue. Here, condensed chromosomes are clearly visible and are separating to form the cores of two new cells.
Related to images 1011, 1012, 1013, 1014, 1015, 1016, 1017, 1018, 1019, and 1021.
Related to images 1011, 1012, 1013, 1014, 1015, 1016, 1017, 1018, 1019, and 1021.
Andrew S. Bajer, University of Oregon, Eugene
View Media
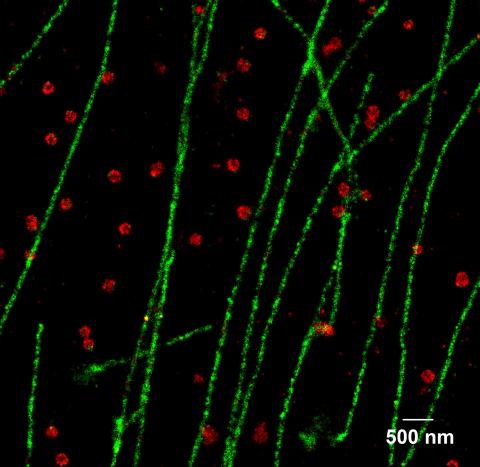
2325: Multicolor STORM
2325: Multicolor STORM
In 2006, scientists developed an optical microscopy technique enabling them to clearly see individual molecules within cells. In 2007, they took the technique, abbreviated STORM, a step further. They identified multicolored probes that let them peer into cells and clearly see multiple cellular components at the same time, such as these microtubules (green) and small hollows called clathrin-coated pits (red). Unlike conventional methods, the multicolor STORM technique produces a crisp and high resolution picture. A sharper view of how cellular components interact will likely help scientists answer some longstanding questions about cell biology.
Xiaowei Zhuang, Harvard University
View Media

6932: Axolotl
6932: Axolotl
An axolotl—a type of salamander—that has been genetically modified so that its developing nervous system glows purple and its Schwann cell nuclei appear light blue. Schwann cells insulate and provide nutrients to peripheral nerve cells. Researchers often study axolotls for their extensive regenerative abilities. They can regrow tails, limbs, spinal cords, brains, and more. The researcher who took this image focuses on the role of the peripheral nervous system during limb regeneration.
This image was captured using a stereo microscope.
Related to images 6927 and 6928.
This image was captured using a stereo microscope.
Related to images 6927 and 6928.
Prayag Murawala, MDI Biological Laboratory and Hannover Medical School.
View Media
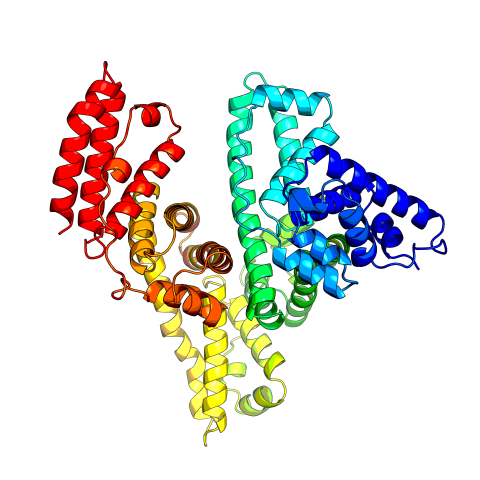
3745: Serum albumin structure 2
3745: Serum albumin structure 2
Serum albumin (SA) is the most abundant protein in the blood plasma of mammals. SA has a characteristic heart-shape structure and is a highly versatile protein. It helps maintain normal water levels in our tissues and carries almost half of all calcium ions in human blood. SA also transports some hormones, nutrients and metals throughout the bloodstream. Despite being very similar to our own SA, those from other animals can cause some mild allergies in people. Therefore, some scientists study SAs from humans and other mammals to learn more about what subtle structural or other differences cause immune responses in the body.
Related to entries 3744 and 3746
Related to entries 3744 and 3746
Wladek Minor, University of Virginia
View Media
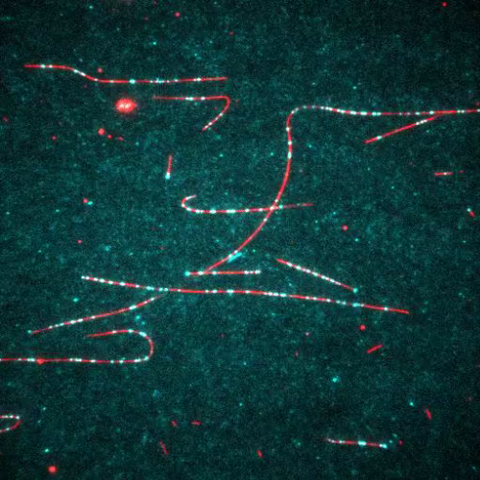
7023: Dynein moving along microtubules
7023: Dynein moving along microtubules
Dynein (green) is a motor protein that “walks” along microtubules (red, part of the cytoskeleton) and carries its cargo along with it. This video was captured through fluorescence microscopy.
Morgan DeSantis, University of Michigan.
View Media

3728: Quorum-sensing inhibitor limits bacterial growth
3728: Quorum-sensing inhibitor limits bacterial growth
To simulate the consequences of disrupting bacterial cell-to-cell communication, called quorum sensing, in the crypts (small chambers within the colon), the researchers experimented with an inhibitor molecule (i.e., antagonist) to turn off quorum sensing in methicillin-resistant Staphylococcus aureus (MRSA), an antibiotic-resistant strain of bacteria that often causes human infections. In this experiment, a medium promoting bacterial growth flows through experimental chambers mimicking the colon environment. The chambers on the right contained no antagonist. In the left chambers, after being added to the flowing medium, the quorum-sensing-inhibiting molecules quickly spread throughout the crevices, inactivating quorum sensing and reducing colonization. These results suggest a potential strategy for addressing MRSA virulence via inhibitors of bacterial communication. You can read more about this research here.
Minyoung Kevin Kim and Bonnie Bassler, Princeton University
View Media
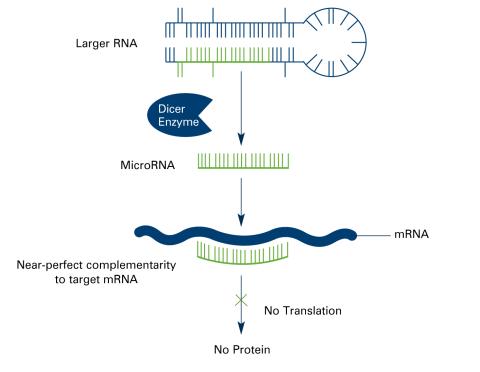
2557: Dicer generates microRNAs (with labels)
2557: Dicer generates microRNAs (with labels)
The enzyme Dicer generates microRNAs by chopping larger RNA molecules into tiny Velcro®-like pieces. MicroRNAs stick to mRNA molecules and prevent the mRNAs from being made into proteins. See image 2556 for an unlabeled version of this illustration. Featured in The New Genetics.
Crabtree + Company
View Media
6520: HeLa cell undergoing division into two daughter cells
6520: HeLa cell undergoing division into two daughter cells
Here, a human HeLa cell (a type of immortal cell line used in laboratory experiments) is undergoing cell division. They come from cervical cancer cells that were obtained in 1951 from Henrietta Lacks, a patient at the Johns Hopkins Hospital. The final stage of division, called cytokinesis, occurs after the genomes—shown in yellow—have split into two new daughter cells. The myosin II is a motor protein shown in blue, and the actin filaments, which are types of protein that support cell structure, are shown in red.
Dylan T. Burnette, Ph.D., Vanderbilt University School of Medicine.
View Media
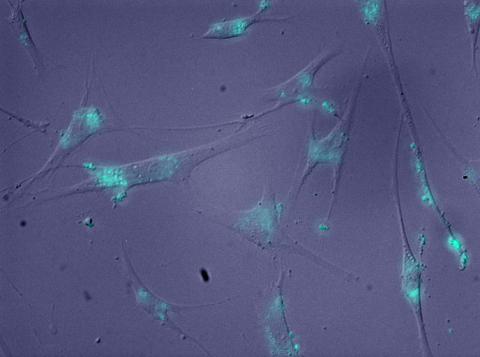
2600: Molecules blocking Huntington's protein production
2600: Molecules blocking Huntington's protein production
The molecules that glow blue in these cultured cells prevent the expression of the mutant proteins that cause Huntington's disease. Biochemist David Corey and others at UT Southwestern Medical Center designed the molecules to specifically target the genetic repeats that code for harmful proteins in people with Huntington's disese. People with Huntington's disease and similar neurodegenerative disorders often have extra copies of a gene segment. Moving from cell cultures to animals will help researchers further explore the potential of their specially crafted molecule to treat brain disorders. In addition to NIGMS, NIH's National Institute of Neurological Disorders and Stroke and National Institute of Biomedical Imaging and Bioengineering also funded this work.
Jiaxin Hu, David W. Dodd and Robert H. E. Hudson, UT Southwestern Medical Center
View Media
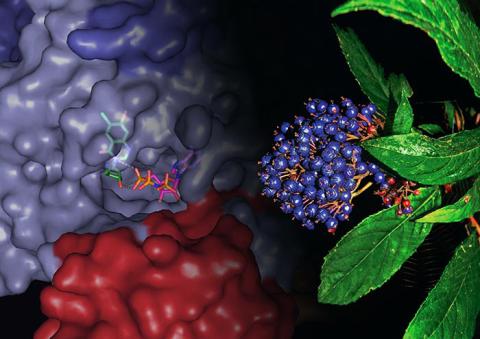
3483: Chang Shan
3483: Chang Shan
For thousands of years, Chinese herbalists have treated malaria using Chang Shan, a root extract from a type of hydrangea that grows in Tibet and Nepal. Recent studies have suggested Chang Shan can also reduce scar formation, treat multiple sclerosis and even slow cancer progression.
Paul Schimmel Lab, Scripps Research Institute
View Media
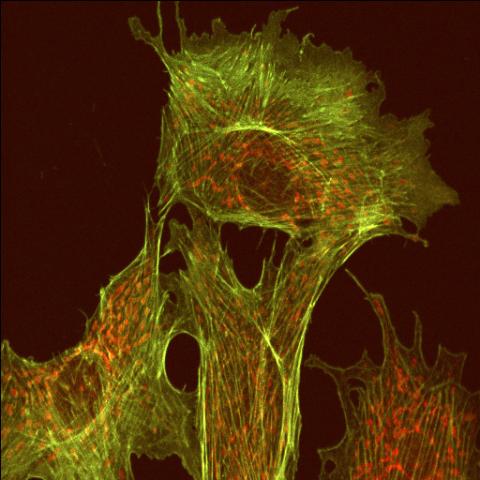
1102: Endothelial cell
1102: Endothelial cell
This image shows two components of the cytoskeleton, microtubules (green) and actin filaments (red), in an endothelial cell derived from a cow lung. The cystoskeleton provides the cell with an inner framework and enables it to move and change shape.
Tina Weatherby Carvalho, University of Hawaii at Manoa
View Media
2693: Fruit fly in the pink
2693: Fruit fly in the pink
Fruit flies are a common model organism for basic medical research.
Crabtree + Company
View Media
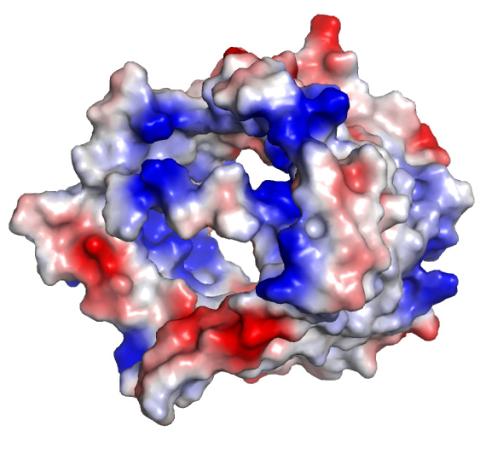
2494: VDAC-1 (3)
2494: VDAC-1 (3)
The structure of the pore-forming protein VDAC-1 from humans. This molecule mediates the flow of products needed for metabolism--in particular the export of ATP--across the outer membrane of mitochondria, the power plants for eukaryotic cells. VDAC-1 is involved in metabolism and the self-destruction of cells--two biological processes central to health.
Related to images 2491, 2495, and 2488.
Related to images 2491, 2495, and 2488.
Gerhard Wagner, Harvard Medical School
View Media
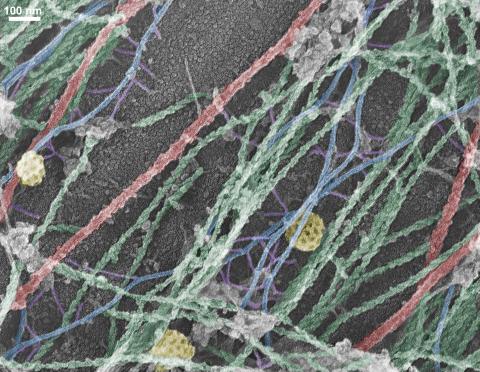
3634: Cells use bubble-like structures called vesicles to transport cargo
3634: Cells use bubble-like structures called vesicles to transport cargo
Cells use bubble-like structures called vesicles (yellow) to import, transport, and export cargo and in cellular communication. A single cell may be filled with thousands of moving vesicles.
This image was part of the Life: Magnified exhibit that ran from June 3, 2014, to January 21, 2015, at Dulles International Airport.
This image was part of the Life: Magnified exhibit that ran from June 3, 2014, to January 21, 2015, at Dulles International Airport.
Tatyana Svitkina, University of Pennsylvania
View Media

6902: Arachnoidiscus diatom
6902: Arachnoidiscus diatom
An Arachnoidiscus diatom with a diameter of 190µm. Diatoms are microscopic algae that have cell walls made of silica, which is the strongest known biological material relative to its density. In Arachnoidiscus, the cell wall is a radially symmetric pillbox-like shell composed of overlapping halves that contain intricate and delicate patterns. Sometimes, Arachnoidiscus is called “a wheel of glass.”
This image was taken with the orientation-independent differential interference contrast microscope.
This image was taken with the orientation-independent differential interference contrast microscope.
Michael Shribak, Marine Biological Laboratory/University of Chicago.
View Media
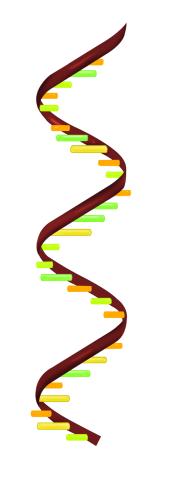
2554: RNA strand
2554: RNA strand
Ribonucleic acid (RNA) has a sugar-phosphate backbone and the bases adenine (A), cytosine (C), guanine (G), and uracil (U). See image 2555 for a labeled version of this illustration. Featured in The New Genetics.
Crabtree + Company
View Media
