Switch to List View
Image and Video Gallery
This is a searchable collection of scientific photos, illustrations, and videos. The images and videos in this gallery are licensed under Creative Commons Attribution Non-Commercial ShareAlike 3.0. This license lets you remix, tweak, and build upon this work non-commercially, as long as you credit and license your new creations under identical terms.
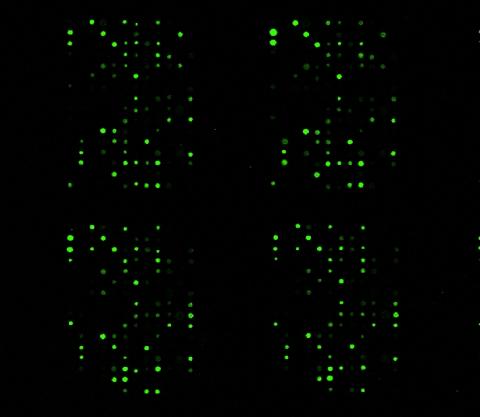
1265: Glycan arrays
1265: Glycan arrays
The signal is obtained by allowing proteins in human serum to interact with glycan (polysaccharide) arrays. The arrays are shown in replicate so the pattern is clear. Each spot contains a specific type of glycan. Proteins have bound to the spots highlighted in green.
Ola Blixt, Scripps Research Institute
View Media
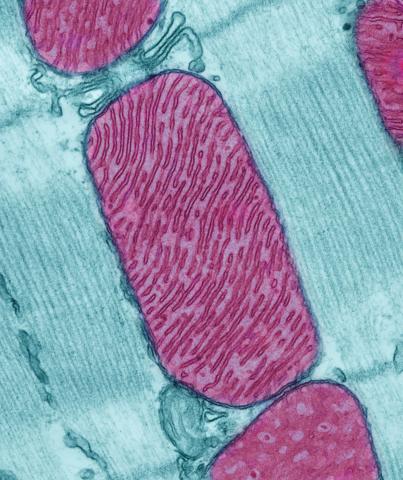
3661: Mitochondria from rat heart muscle cell
3661: Mitochondria from rat heart muscle cell
These mitochondria (red) are from the heart muscle cell of a rat. Mitochondria have an inner membrane that folds in many places (and that appears here as striations). This folding vastly increases the surface area for energy production. Nearly all our cells have mitochondria. Related to image 3664.
National Center for Microscopy and Imaging Research
View Media
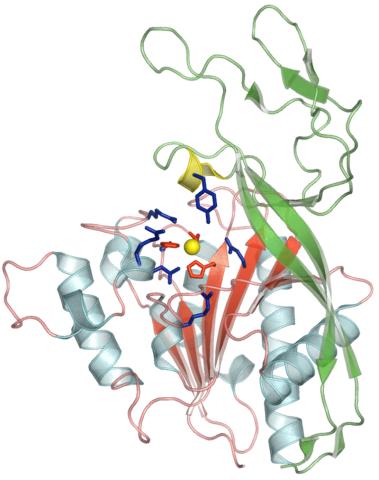
2352: Human aspartoacylase
2352: Human aspartoacylase
Model of aspartoacylase, a human enzyme involved in brain metabolism.
Center for Eukaryotic Structural Genomics, PSI
View Media
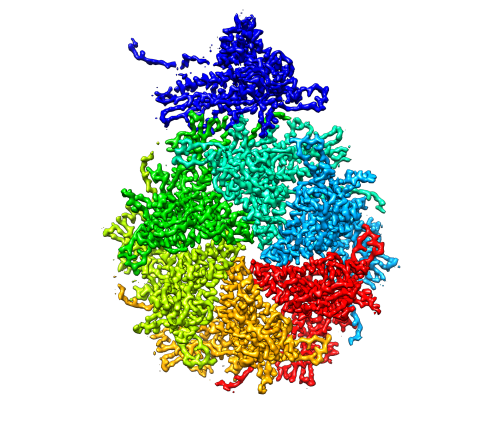
5875: Bacteriophage P22 capsid, detail
5875: Bacteriophage P22 capsid, detail
Detail of a subunit of the capsid, or outer cover, of bacteriophage P22, a virus that infects the Salmonella bacteria. Cryo-electron microscopy (cryo-EM) was used to capture details of the capsid proteins, each shown here in a separate color. Thousands of cryo-EM scans capture the structure and shape of all the individual proteins in the capsid and their position relative to other proteins. A computer model combines these scans into the image shown here. Related to image 5874.
Dr. Wah Chiu, Baylor College of Medicine
View Media
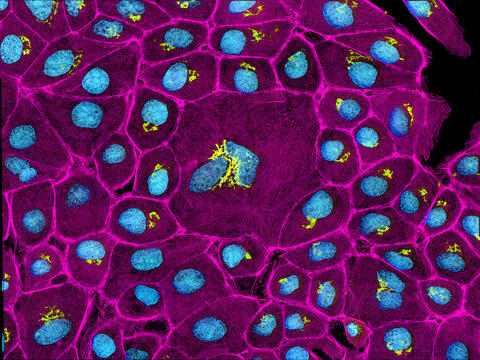
3647: Epithelial cells
3647: Epithelial cells
This image mostly shows normal cultured epithelial cells expressing green fluorescent protein targeted to the Golgi apparatus (yellow-green) and stained for actin (magenta) and DNA (cyan). The middle cell is an abnormal large multinucleated cell. All the cells in this image have a Golgi but not all are expressing the targeted recombinant fluorescent protein.
Tom Deerinck, National Center for Microscopy and Imaging Research (NCMIR)
View Media

2371: NMR spectrometer
2371: NMR spectrometer
This photo shows a Varian Unity Inova 900 MHz, 21.1 T standard bore magnet Nuclear Magnetic Resonnance (NMR) spectrometer. NMR spectroscopy provides data used to determine the structures of proteins in solution, rather than in crystal form, as in X-ray crystallography. The technique is limited to smaller proteins or protein fragments in a high throughput approach.
Center for Eukaryotic Structural Genomics
View Media
2740: Early life of a protein
2740: Early life of a protein
This illustration represents the early life of a protein—specifically, apomyoglobin—as it is synthesized by a ribosome and emerges from the ribosomal tunnel, which contains the newly formed protein's conformation. The synthesis occurs in the complex swirl of the cell medium, filled with interactions among many molecules. Researchers in Silvia Cavagnero's laboratory are studying the structure and dynamics of newly made proteins and polypeptides using spectroscopic and biochemical techniques.
Silvia Cavagnero, University of Wisconsin, Madison
View Media
6547: Cell Nucleus and Lipid Droplets
6547: Cell Nucleus and Lipid Droplets
A cell nucleus (blue) surrounded by lipid droplets (yellow). Exogenously expressed, S-tagged UBXD8 (green) recruits endogenous p97/VCP (red) to the surface of lipid droplets in oleate-treated HeLa cells. Nucleus stained with DAPI.
James Olzmann, University of California, Berkeley
View Media
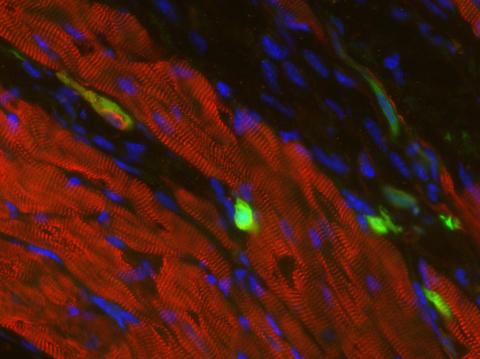
3273: Heart muscle with reprogrammed skin cells
3273: Heart muscle with reprogrammed skin cells
Skins cells were reprogrammed into heart muscle cells. The cells highlighted in green are remaining skin cells. Red indicates a protein that is unique to heart muscle. The technique used to reprogram the skin cells into heart cells could one day be used to mend heart muscle damaged by disease or heart attack. Image and caption information courtesy of the California Institute for Regenerative Medicine.
Deepak Srivastava, Gladstone Institute of Cardiovascular Disease, via CIRM
View Media
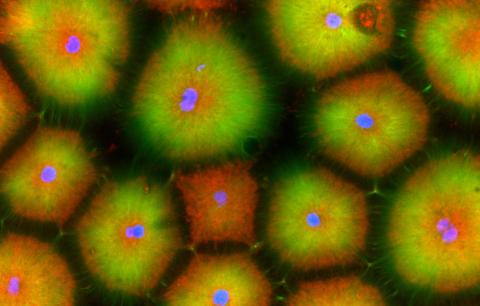
6592: Cell-like compartments from frog eggs 5
6592: Cell-like compartments from frog eggs 5
Cell-like compartments that spontaneously emerged from scrambled frog eggs, with nuclei (blue) from frog sperm. Endoplasmic reticulum (red) and microtubules (green) are also visible. Image created using confocal microscopy.
For more photos of cell-like compartments from frog eggs view: 6584, 6585, 6586, 6591, and 6593.
For videos of cell-like compartments from frog eggs view: 6587, 6588, 6589, and 6590.
Xianrui Cheng, Stanford University School of Medicine.
View Media

7012: Adult Hawaiian bobtail squid burying in the sand
7012: Adult Hawaiian bobtail squid burying in the sand
Each morning, the nocturnal Hawaiian bobtail squid, Euprymna scolopes, hides from predators by digging into the sand. At dusk, it leaves the sand again to hunt.
Related to image 7010 and 7011.
Related to image 7010 and 7011.
Margaret J. McFall-Ngai, Carnegie Institution for Science/California Institute of Technology, and Edward G. Ruby, California Institute of Technology.
View Media
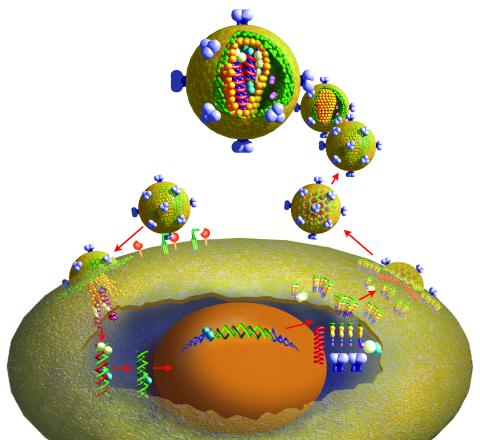
2513: Life of an AIDS virus
2513: Life of an AIDS virus
HIV is a retrovirus, a type of virus that carries its genetic material not as DNA but as RNA. Long before anyone had heard of HIV, researchers in labs all over the world studied retroviruses, tracing out their life cycle and identifying the key proteins the viruses use to infect cells. When HIV was identified as a retrovirus, these studies gave AIDS researchers an immediate jump-start. The previously identified viral proteins became initial drug targets. See images 2514 and 2515 for labeled versions of this illustration. Featured in The Structures of Life.
Crabtree + Company
View Media
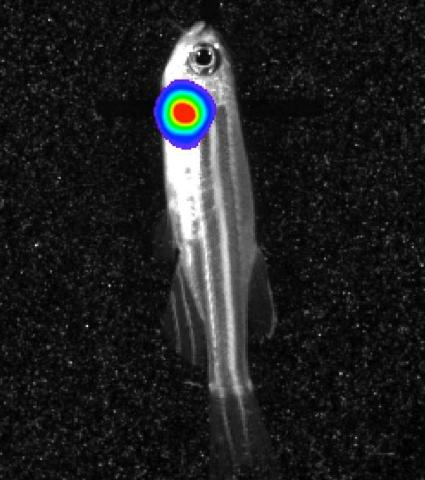
3558: Bioluminescent imaging in adult zebrafish - lateral view
3558: Bioluminescent imaging in adult zebrafish - lateral view
Luciferase-based imaging enables visualization and quantification of internal organs and transplanted cells in live adult zebrafish. In this image, a cardiac muscle-restricted promoter drives firefly luciferase expression (lateral view).
For imagery of both the lateral and overhead view go to 3556.
For imagery of the overhead view go to 3557.
For more information about the illumated area go to 3559.
For imagery of both the lateral and overhead view go to 3556.
For imagery of the overhead view go to 3557.
For more information about the illumated area go to 3559.
Kenneth Poss, Duke University
View Media
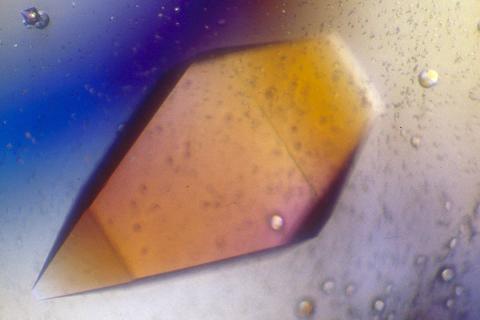
2400: Pig trypsin (1)
2400: Pig trypsin (1)
A crystal of porcine trypsin protein created for X-ray crystallography, which can reveal detailed, three-dimensional protein structures.
Alex McPherson, University of California, Irvine
View Media
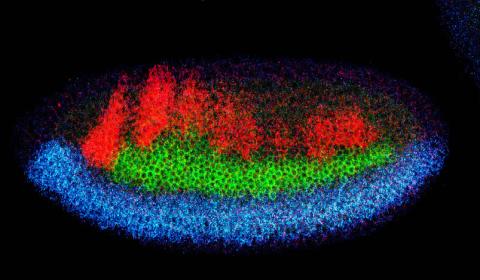
2327: Neural development
2327: Neural development
Using techniques that took 4 years to design, a team of developmental biologists showed that certain proteins can direct the subdivision of fruit fly and chicken nervous system tissue into the regions depicted here in blue, green, and red. Molecules called bone morphogenetic proteins (BMPs) helped form this fruit fly embryo. While scientists knew that BMPs play a major role earlier in embryonic development, they didn't know how the proteins help organize nervous tissue. The findings suggest that BMPs are part of an evolutionarily conserved mechanism for organizing the nervous system. The National Institute of Neurological Disorders and Stroke also supported this work.
Mieko Mizutani and Ethan Bier, University of California, San Diego, and Henk Roelink, University of Washington
View Media
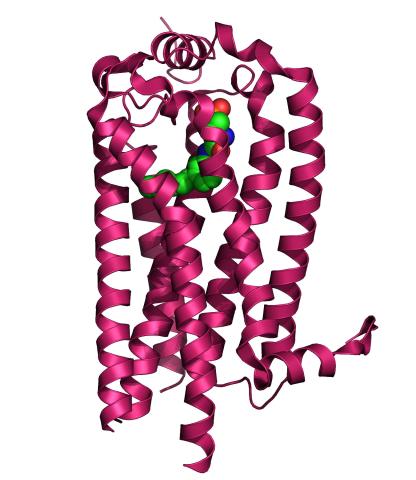
3362: Sphingolipid S1P1 receptor
3362: Sphingolipid S1P1 receptor
The receptor is shown bound to an antagonist, ML056.
Raymond Stevens, The Scripps Research Institute
View Media

2356: Student overseeing protein cloning robot
2356: Student overseeing protein cloning robot
Student Christina Hueneke of the Midwest Center for Structural Genomics is overseeing a protein cloning robot. The robot was designed as part of an effort to exponentially increase the output of a traditional wet lab. Part of the center's goal is to cut the average cost of analyzing a protein from $200,000 to $20,000 and to slash the average time from months to days and hours.
Midwest Center for Structural Genomics
View Media
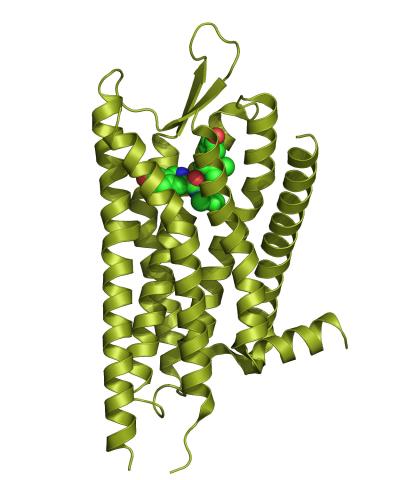
3359: Kappa opioid receptor
3359: Kappa opioid receptor
The receptor is shown bound to an antagonist, JDTic.
Raymond Stevens, The Scripps Research Institute
View Media

6887: Chromatin in human fibroblast
6887: Chromatin in human fibroblast
The nucleus of a human fibroblast cell with chromatin—a substance made up of DNA and proteins—shown in various colors. Fibroblasts are one of the most common types of cells in mammalian connective tissue, and they play a key role in wound healing and tissue repair. This image was captured using Stochastic Optical Reconstruction Microscopy (STORM).
Related to images 6888 and 6893.
Related to images 6888 and 6893.
Melike Lakadamyali, Perelman School of Medicine at the University of Pennsylvania.
View Media
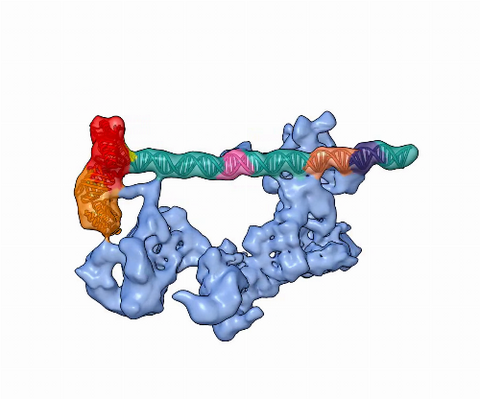
5730: Dynamic cryo-EM model of the human transcription preinitiation complex
5730: Dynamic cryo-EM model of the human transcription preinitiation complex
Gene transcription is a process by which information encoded in DNA is transcribed into RNA. It's essential for all life and requires the activity of proteins, called transcription factors, that detect where in a DNA strand transcription should start. In eukaryotes (i.e., those that have a nucleus and mitochondria), a protein complex comprising 14 different proteins is responsible for sniffing out transcription start sites and starting the process. This complex represents the core machinery to which an enzyme, named RNA polymerase, can bind to and read the DNA and transcribe it to RNA. Scientists have used cryo-electron microscopy (cryo-EM) to visualize the TFIID-RNA polymerase-DNA complex in unprecedented detail. This animation shows the different TFIID components as they contact DNA and recruit the RNA polymerase for gene transcription.
To learn more about the research that has shed new light on gene transcription, see this news release from Berkeley Lab.
Related to image 3766.
To learn more about the research that has shed new light on gene transcription, see this news release from Berkeley Lab.
Related to image 3766.
Eva Nogales, Berkeley Lab
View Media
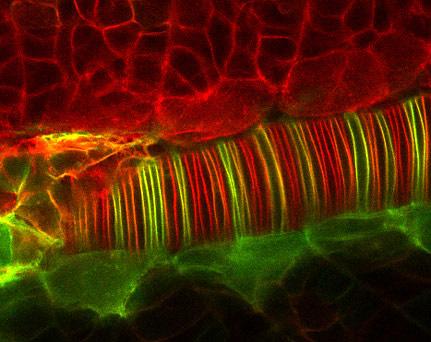
2328: Neural tube development
2328: Neural tube development
Proteins in the neural tissues of this zebrafish embryo direct cells to line up and form the neural tube, which will become the spinal cord and brain. Studies of zebrafish embryonic development may help pinpoint the underlying cause of common neural tube defects--such as spina bifida--which occur in about 1 in 1,000 newborn children.
Alexander Schier, Harvard University
View Media
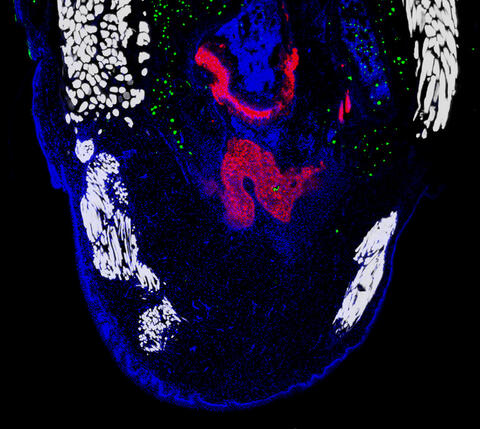
6968: Regenerating lizard tail
6968: Regenerating lizard tail
The interior of a regenerating lizard tail 14 days after the original tail was amputated. Cell nuclei (blue), proliferating cells (green), cartilage (red), and muscle (white) have been visualized with immunofluorescence staining.
Thomas Lozito, University of Southern California.
View Media
6597: Pathways – Bacteria vs. Viruses: What's the Difference?
6597: Pathways – Bacteria vs. Viruses: What's the Difference?
Learn about how bacteria and viruses differ, how they each can make you sick, and how they can or cannot be treated. Discover more resources from NIGMS’ Pathways collaboration with Scholastic. View the video on YouTube for closed captioning.
National Institute of General Medical Sciences
View Media
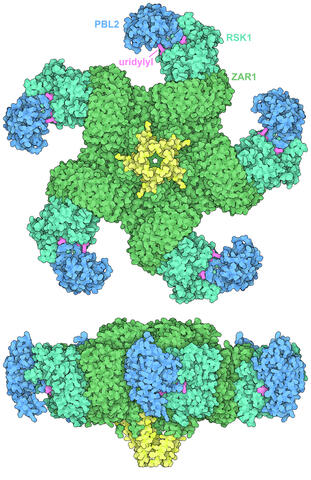
7002: Plant resistosome
7002: Plant resistosome
The research organism Arabidopsis thaliana forms a large molecular machine called a resistosome to fight off infections. This illustration shows the top and side views of the fully-formed resistosome assembly (PDB entry 6J5T), composed of different proteins including one the plant uses as a decoy, PBL2 (dark blue), that gets uridylylated to begin the process of building the resistosome (uridylyl groups in magenta). Other proteins include RSK1 (turquoise) and ZAR1 (green) subunits. The ends of the ZAR1 subunits (yellow) form a funnel-like protrusion on one side of the assembly (seen in the side view). The funnel can carry out the critical protective function of the resistosome by inserting itself into the cell membrane to form a pore, which leads to a localized programmed cell death. The death of the infected cell helps protect the rest of the plant.
Amy Wu and Christine Zardecki, RCSB Protein Data Bank.
View Media
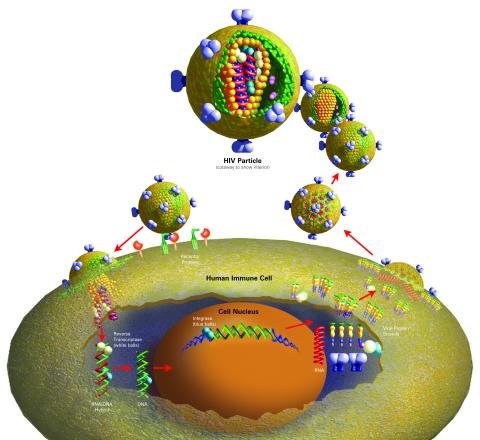
2514: Life of an AIDS virus (with labels)
2514: Life of an AIDS virus (with labels)
HIV is a retrovirus, a type of virus that carries its genetic material not as DNA but as RNA. Long before anyone had heard of HIV, researchers in labs all over the world studied retroviruses, tracing out their life cycle and identifying the key proteins the viruses use to infect cells. When HIV was identified as a retrovirus, these studies gave AIDS researchers an immediate jump-start. The previously identified viral proteins became initial drug targets. See images 2513 and 2515 for other versions of this illustration. Featured in The Structures of Life.
Crabtree + Company
View Media
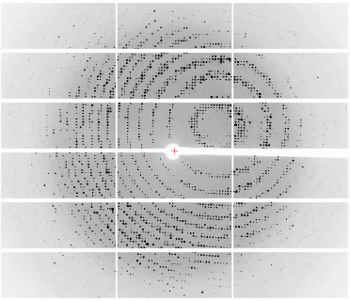
6765: X-ray diffraction pattern from a crystallized cefotaxime-CCD-1 complex
6765: X-ray diffraction pattern from a crystallized cefotaxime-CCD-1 complex
CCD-1 is an enzyme produced by the bacterium Clostridioides difficile that helps it resist antibiotics. Researchers crystallized complexes where a CCD-1 molecule and a molecule of the antibiotic cefotaxime were bound together. Then, they shot X-rays at the complexes to determine their structure—a process known as X-ray crystallography. This image shows the X-ray diffraction pattern of a complex.
Related to images 6764, 6766, and 6767.
Related to images 6764, 6766, and 6767.
Keith Hodgson, Stanford University.
View Media
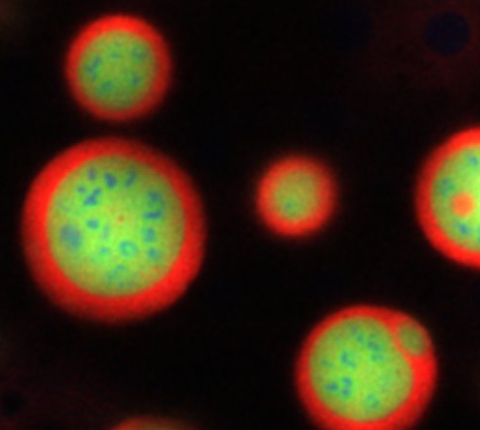
3793: Nucleolus subcompartments spontaneously self-assemble 4
3793: Nucleolus subcompartments spontaneously self-assemble 4
What looks a little like distant planets with some mysterious surface features are actually assemblies of proteins normally found in the cell's nucleolus, a small but very important protein complex located in the cell's nucleus. It forms on the chromosomes at the location where the genes for the RNAs are that make up the structure of the ribosome, the indispensable cellular machine that makes proteins from messenger RNAs.
However, how the nucleolus grows and maintains its structure has puzzled scientists for some time. It turns out that even though it looks like a simple liquid blob, it's rather well-organized, consisting of three distinct layers: the fibrillar center, where the RNA polymerase is active; the dense fibrillar component, which is enriched in the protein fibrillarin; and the granular component, which contains a protein called nucleophosmin. Researchers have now discovered that this multilayer structure of the nucleolus arises from differences in how the proteins in each compartment mix with water and with each other. These differences let the proteins readily separate from each other into the three nucleolus compartments.
This photo of nucleolus proteins in the eggs of a commonly used lab animal, the frog Xenopus laevis, shows each of the nucleolus compartments (the granular component is shown in red, the fibrillarin in yellow-green, and the fibrillar center in blue). The researchers have found that these compartments spontaneously fuse with each other on encounter without mixing with the other compartments.
For more details on this research, see this press release from Princeton. Related to video 3789, video 3791 and image 3792.
However, how the nucleolus grows and maintains its structure has puzzled scientists for some time. It turns out that even though it looks like a simple liquid blob, it's rather well-organized, consisting of three distinct layers: the fibrillar center, where the RNA polymerase is active; the dense fibrillar component, which is enriched in the protein fibrillarin; and the granular component, which contains a protein called nucleophosmin. Researchers have now discovered that this multilayer structure of the nucleolus arises from differences in how the proteins in each compartment mix with water and with each other. These differences let the proteins readily separate from each other into the three nucleolus compartments.
This photo of nucleolus proteins in the eggs of a commonly used lab animal, the frog Xenopus laevis, shows each of the nucleolus compartments (the granular component is shown in red, the fibrillarin in yellow-green, and the fibrillar center in blue). The researchers have found that these compartments spontaneously fuse with each other on encounter without mixing with the other compartments.
For more details on this research, see this press release from Princeton. Related to video 3789, video 3791 and image 3792.
Nilesh Vaidya, Princeton University
View Media
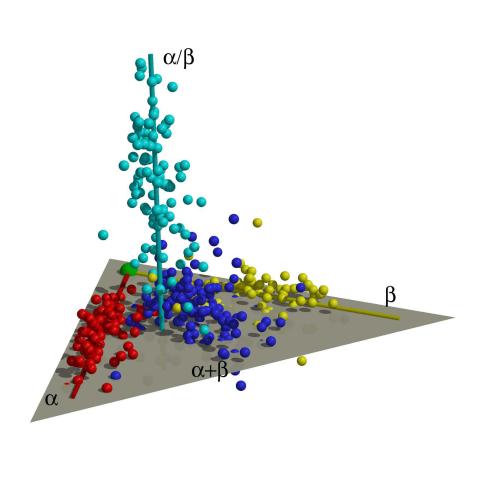
2365: Map of protein structures 01
2365: Map of protein structures 01
A global "map of the protein structure universe." The Berkeley Structural Genomics Center has developed a method to visualize the vast universe of protein structures in which proteins of similar structure are located close together and those of different structures far away in the space. This map, constructed using about 500 of the most common protein folds, reveals a highly non-uniform distribution, and shows segregation between four elongated regions corresponding to four different protein classes (shown in four different colors). Such a representation reveals a high-level of organization of the protein structure universe.
Berkeley Structural Genomics Center, PSI
View Media
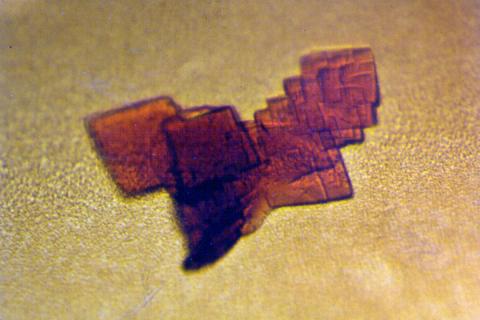
2392: Sheep hemoglobin crystal
2392: Sheep hemoglobin crystal
A crystal of sheep hemoglobin protein created for X-ray crystallography, which can reveal detailed, three-dimensional protein structures.
Alex McPherson, University of California, Irvine
View Media
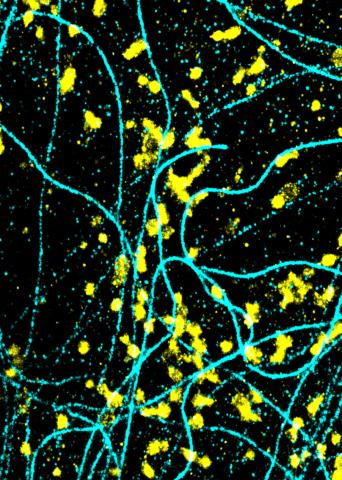
6889: Lysosomes and microtubules
6889: Lysosomes and microtubules
Lysosomes (yellow) and detyrosinated microtubules (light blue). Lysosomes are bubblelike organelles that take in molecules and use enzymes to break them down. Microtubules are strong, hollow fibers that provide structural support to cells. The researchers who took this image found that in epithelial cells, detyrosinated microtubules are a small subset of fibers, and they concentrate lysosomes around themselves. This image was captured using Stochastic Optical Reconstruction Microscopy (STORM).
Related to images 6890, 6891, and 6892.
Related to images 6890, 6891, and 6892.
Melike Lakadamyali, Perelman School of Medicine at the University of Pennsylvania.
View Media
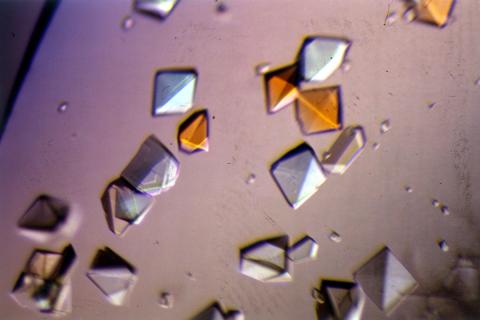
2412: Pig alpha amylase
2412: Pig alpha amylase
Crystals of porcine alpha amylase protein created for X-ray crystallography, which can reveal detailed, three-dimensional protein structures.
Alex McPherson, University of California, Irvine
View Media
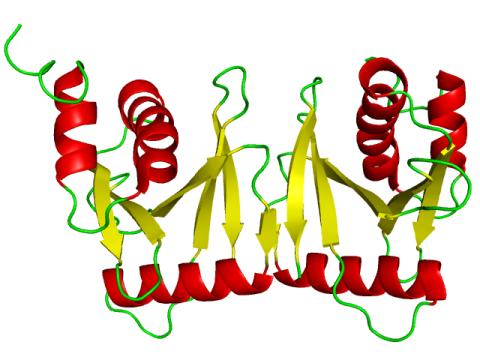
2351: tRNA splicing enzyme endonuclease in humans
2351: tRNA splicing enzyme endonuclease in humans
An NMR solution structure model of the transfer RNA splicing enzyme endonuclease in humans (subunit Sen15). This represents the first structure of a eukaryotic tRNA splicing endonuclease subunit.
Center for Eukaryotic Structural Genomics, PSI
View Media
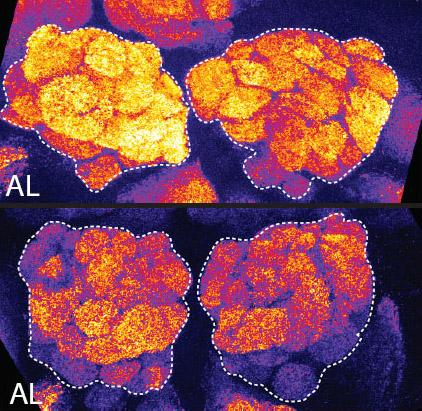
2596: Sleep and the fly brain
2596: Sleep and the fly brain
In the top snapshots, the brain of a sleep-deprived fruit fly glows orange, marking high concentrations of a synaptic protein called Bruchpilot (BRP) involved in communication between neurons. The color particularly lights up brain areas associated with learning. By contrast, the bottom images from a well-rested fly show lower levels of the protein. These pictures illustrate the results of an April 2009 study showing that sleep reduces the protein's levels, suggesting that such "downscaling" resets the brain to normal levels of synaptic activity and makes it ready to learn after a restful night.
Chiara Cirelli, University of Wisconsin-Madison
View Media
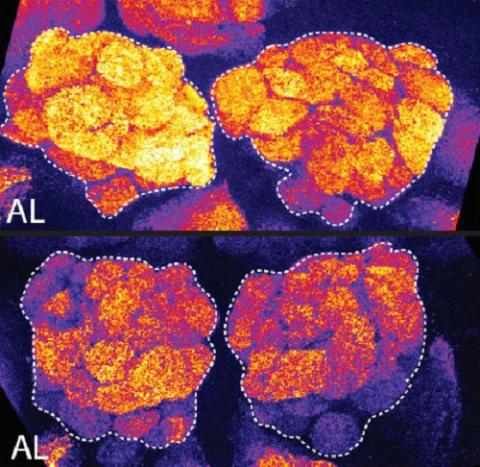
3490: Brains of sleep-deprived and well-rested fruit flies
3490: Brains of sleep-deprived and well-rested fruit flies
On top, the brain of a sleep-deprived fly glows orange because of Bruchpilot, a communication protein between brain cells. These bright orange brain areas are associated with learning. On the bottom, a well-rested fly shows lower levels of Bruchpilot, which might make the fly ready to learn after a good night's rest.
Chiara Cirelli, University of Wisconsin-Madison
View Media
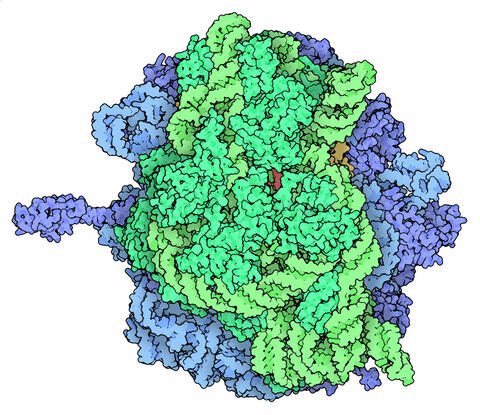
5780: Ribosome illustration from PDB
5780: Ribosome illustration from PDB
Ribosomes are complex machines made up of more than 50 proteins and three or four strands of genetic material called ribosomal RNA (rRNA). The busy cellular machines make proteins, which are critical to almost every structure and function in the cell. To do so, they read protein-building instructions, which come as strands of messenger RNA. Ribosomes are found in all forms of cellular life—people, plants, animals, even bacteria. This illustration of a bacterial ribosome was produced using detailed information about the position of every atom in the complex. Several antibiotic medicines work by disrupting bacterial ribosomes but leaving human ribosomes alone. Scientists are carefully comparing human and bacterial ribosomes to spot differences between the two. Structures that are present only in the bacterial version could serve as targets for new antibiotic medications.
From PDB’s Molecule of the Month collection (direct link: http://pdb101.rcsb.org/motm/121) Molecule of the Month illustrations are available under a CC-BY-4.0 license. Attribution should be given to David S. Goodsell and the RCSB PDB.
View Media

3772: The Proteasome: The Cell's Trash Processor in Action
3772: The Proteasome: The Cell's Trash Processor in Action
Our cells are constantly removing and recycling molecular waste. This video shows one way cells process their trash.
View Media
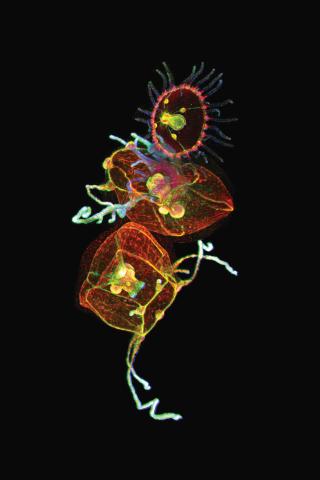
3636: Jellyfish, viewed with ZEISS Lightsheet Z.1 microscope
3636: Jellyfish, viewed with ZEISS Lightsheet Z.1 microscope
Jellyfish are especially good models for studying the evolution of embryonic tissue layers. Despite being primitive, jellyfish have a nervous system (stained green here) and musculature (red). Cell nuclei are stained blue. By studying how tissues are distributed in this simple organism, scientists can learn about the evolution of the shapes and features of diverse animals.
This image was part of the Life: Magnified exhibit that ran from June 3, 2014, to January 21, 2015, at Dulles International Airport.
This image was part of the Life: Magnified exhibit that ran from June 3, 2014, to January 21, 2015, at Dulles International Airport.
Helena Parra, Pompeu Fabra University, Spain
View Media
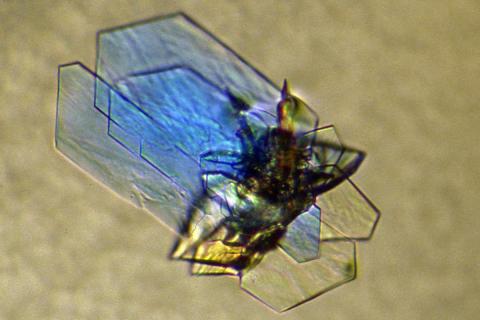
2410: DNase
2410: DNase
Crystals of DNase protein created for X-ray crystallography, which can reveal detailed, three-dimensional protein structures.
Alex McPherson, University of California, Irvine
View Media
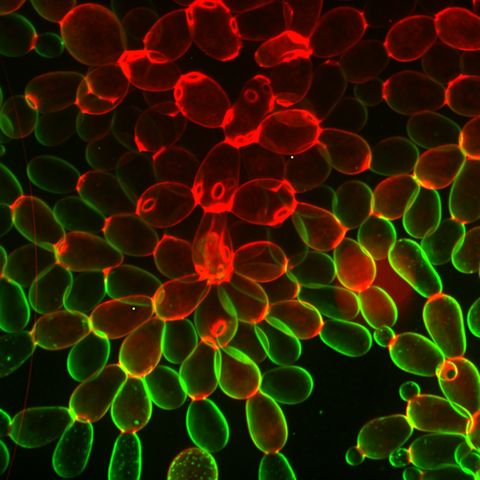
6969: Snowflake yeast 1
6969: Snowflake yeast 1
Multicellular yeast called snowflake yeast that researchers created through many generations of directed evolution from unicellular yeast. Stained cell membranes (green) and cell walls (red) reveal the connections between cells. Younger cells take up more cell membrane stain, while older cells take up more cell wall stain, leading to the color differences seen here. This image was captured using spinning disk confocal microscopy.
Related to images 6970 and 6971.
Related to images 6970 and 6971.
William Ratcliff, Georgia Institute of Technology.
View Media
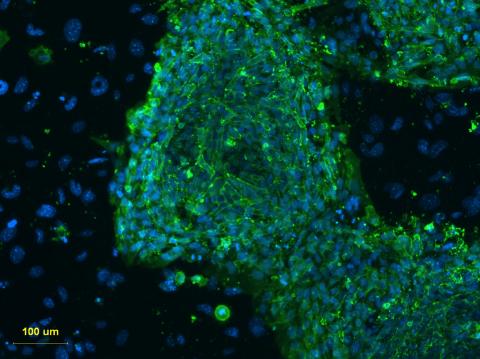
3274: Human embryonic stem cells on feeder cells
3274: Human embryonic stem cells on feeder cells
This fluorescent microscope image shows human embryonic stem cells whose nuclei are stained green. Blue staining shows the surrounding supportive feeder cells. Image and caption information courtesy of the California Institute for Regenerative Medicine. See related image 3275.
Michael Longaker lab, Stanford University School of Medicine, via CIRM
View Media
2430: Fruit fly retina 01
2430: Fruit fly retina 01
Image showing rhabdomeres (red), the light-sensitive structures in the fruit fly retina, and rhodopsin-4 (blue), a light-sensing molecule.
Hermann Steller, Rockefeller University
View Media
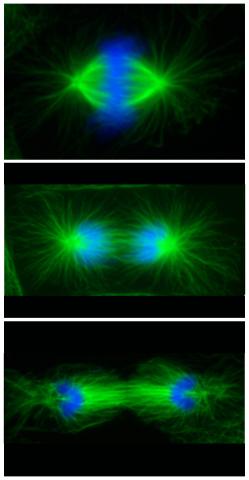
3442: Cell division phases in Xenopus frog cells
3442: Cell division phases in Xenopus frog cells
These images show three stages of cell division in Xenopus XL177 cells, which are derived from tadpole epithelial cells. They are (from top): metaphase, anaphase and telophase. The microtubules are green and the chromosomes are blue. Related to 3443.
Claire Walczak, who took them while working as a postdoc in the laboratory of Timothy Mitchison
View Media

1283: Vesicle traffic
1283: Vesicle traffic
This illustration shows vesicle traffic inside a cell. The double membrane that bounds the nucleus flows into the ribosome-studded rough endoplasmic reticulum (purple), where membrane-embedded proteins are manufactured. Proteins are processed and lipids are manufactured in the smooth endoplasmic reticulum (blue) and Golgi apparatus (green). Vesicles that fuse with the cell membrane release their contents outside the cell. The cell can also take in material from outside by having vesicles pinch off from the cell membrane.
Judith Stoffer
View Media
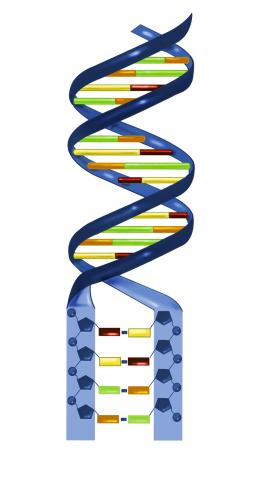
2541: Nucleotides make up DNA
2541: Nucleotides make up DNA
DNA consists of two long, twisted chains made up of nucleotides. Each nucleotide contains one base, one phosphate molecule, and the sugar molecule deoxyribose. The bases in DNA nucleotides are adenine, thymine, cytosine, and guanine. See image 2542 for a labeled version of this illustration. Featured in The New Genetics.
Crabtree + Company
View Media
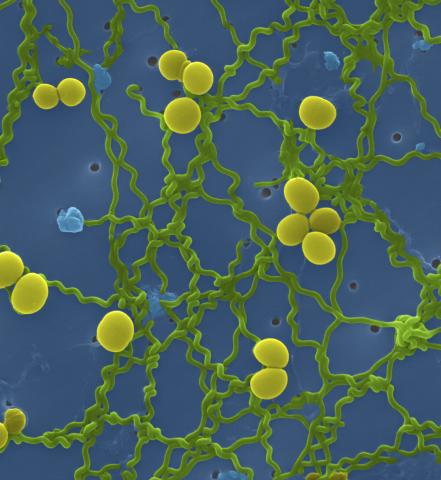
1166: Leptospira bacteria
1166: Leptospira bacteria
Leptospira, shown here in green, is a type (genus) of elongated, spiral-shaped bacteria. Infection can cause Weil's disease, a kind of jaundice, in humans.
Tina Weatherby Carvalho, University of Hawaii at Manoa
View Media
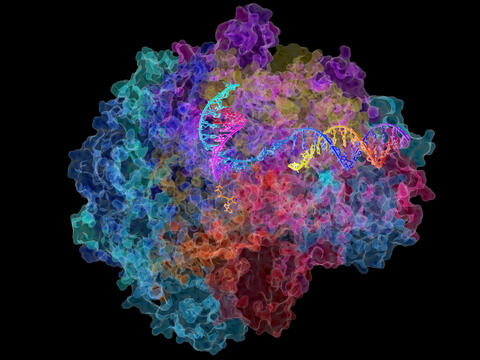
2484: RNA Polymerase II
2484: RNA Polymerase II
NIGMS-funded researchers led by Roger Kornberg solved the structure of RNA polymerase II. This is the enzyme in mammalian cells that catalyzes the transcription of DNA into messenger RNA, the molecule that in turn dictates the order of amino acids in proteins. For his work on the mechanisms of mammalian transcription, Kornberg received the Nobel Prize in Chemistry in 2006.
David Bushnell, Ken Westover and Roger Kornberg, Stanford University
View Media
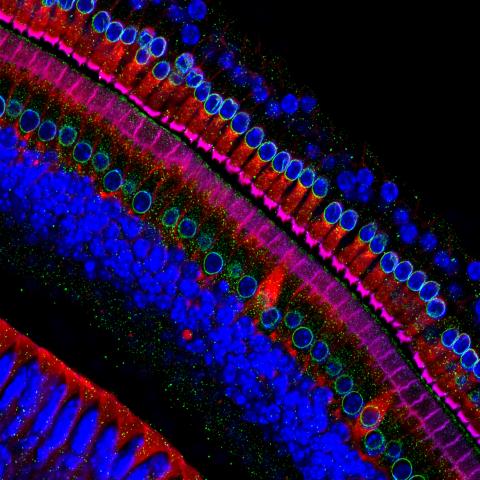
3618: Hair cells: the sound-sensing cells in the ear
3618: Hair cells: the sound-sensing cells in the ear
These cells get their name from the hairlike structures that extend from them into the fluid-filled tube of the inner ear. When sound reaches the ear, the hairs bend and the cells convert this movement into signals that are relayed to the brain. When we pump up the music in our cars or join tens of thousands of cheering fans at a football stadium, the noise can make the hairs bend so far that they actually break, resulting in long-term hearing loss.
This image was part of the Life: Magnified exhibit that ran from June 3, 2014, to January 21, 2015, at Dulles International Airport.
This image was part of the Life: Magnified exhibit that ran from June 3, 2014, to January 21, 2015, at Dulles International Airport.
Henning Horn, Brian Burke, and Colin Stewart, Institute of Medical Biology, Agency for Science, Technology, and Research, Singapore
View Media
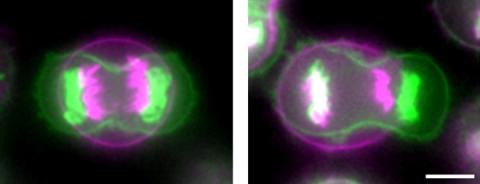
3648: Symmetrically and asymmetrically elongating cells
3648: Symmetrically and asymmetrically elongating cells
Merged fluorescent images of symmetrically (left) or asymmetrically (right) elongating HeLa cells at the end of early anaphase (magenta) and late anaphase (green). Chromosomes and cortical actin are visualized by expressing mCherry-histone H2B and Lifeact-mCherry. Scale bar, 10µm. See the PubMed abstract of this research.
Tomomi Kiyomitsu and Iain M. Cheeseman, Whitehead Institute for Biomedical Research
View Media
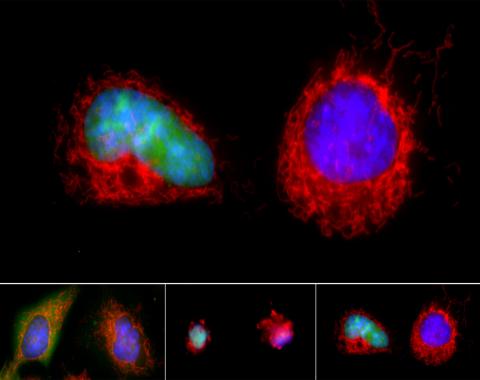
3486: Apoptosis reversed
3486: Apoptosis reversed
Two healthy cells (bottom, left) enter into apoptosis (bottom, center) but spring back to life after a fatal toxin is removed (bottom, right; top).
Hogan Tang of the Denise Montell Lab, Johns Hopkins University School of Medicine
View Media
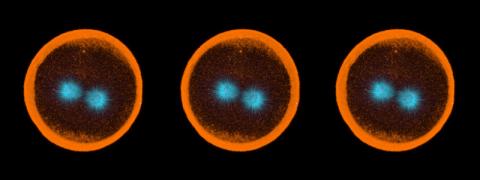
1050: Sea urchin embryo 04
1050: Sea urchin embryo 04
Stereo triplet of a sea urchin embryo stained to reveal actin filaments (orange) and microtubules (blue). This image is part of a series of images: image 1047, image 1048, image 1049, image 1051 and image 1052.
George von Dassow, University of Washington
View Media
