Switch to List View
Image and Video Gallery
This is a searchable collection of scientific photos, illustrations, and videos. The images and videos in this gallery are licensed under Creative Commons Attribution Non-Commercial ShareAlike 3.0. This license lets you remix, tweak, and build upon this work non-commercially, as long as you credit and license your new creations under identical terms.
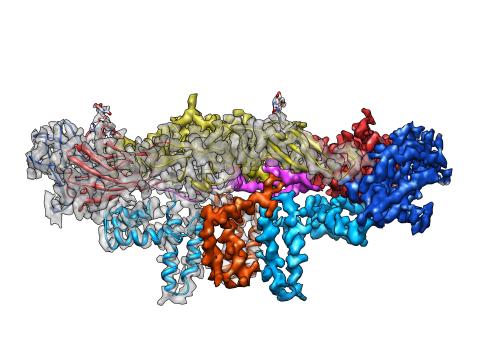
3758: Dengue virus membrane protein structure
3758: Dengue virus membrane protein structure
Dengue virus is a mosquito-borne illness that infects millions of people in the tropics and subtropics each year. Like many viruses, dengue is enclosed by a protective membrane. The proteins that span this membrane play an important role in the life cycle of the virus. Scientists used cryo-EM to determine the structure of a dengue virus at a 3.5-angstrom resolution to reveal how the membrane proteins undergo major structural changes as the virus matures and infects a host. The image shows a side view of the structure of a protein composed of two smaller proteins, called E and M. Each E and M contributes two molecules to the overall protein structure (called a heterotetramer), which is important for assembling and holding together the viral membrane, i.e., the shell that surrounds the genetic material of the dengue virus. The dengue protein's structure has revealed some portions in the protein that might be good targets for developing medications that could be used to combat dengue virus infections. For more on cryo-EM see the blog post Cryo-Electron Microscopy Reveals Molecules in Ever Greater Detail. You can watch a rotating view of the dengue virus surface structure in video 3748.
Hong Zhou, UCLA
View Media

2326: Nano-rainbow
2326: Nano-rainbow
These vials may look like they're filled with colored water, but they really contain nanocrystals reflecting different colors under ultraviolet light. The tiny crystals, made of semiconducting compounds, are called quantum dots. Depending on their size, the dots emit different colors that let scientists use them as a tool for detecting particular genes, proteins, and other biological molecules.
Shuming Nie, Emory University
View Media
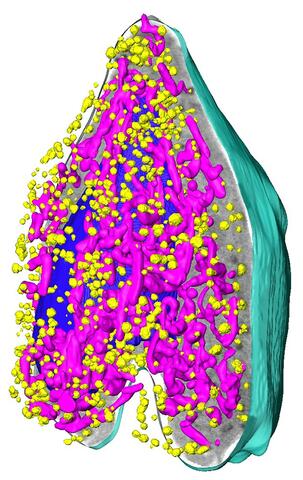
6605: Soft X-ray tomography of a pancreatic beta cell
6605: Soft X-ray tomography of a pancreatic beta cell
A color-coded, 3D model of a rat pancreatic β cell. This type of cell produces insulin, a hormone that helps regulate blood sugar. Visible are mitochondria (pink), insulin vesicles (yellow), the nucleus (dark blue), and the plasma membrane (teal). This model was created based on soft X-ray tomography (SXT) images.
Carolyn Larabell, University of California, San Francisco.
View Media
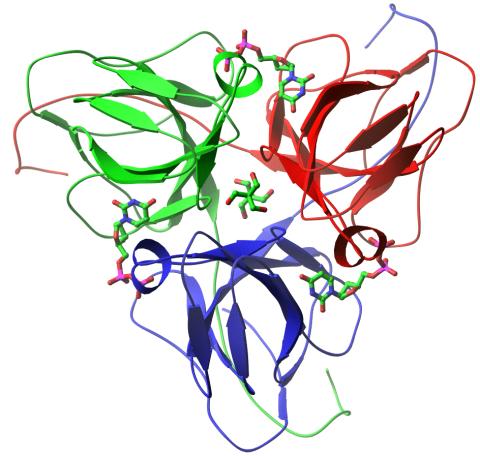
2381: dUTP pyrophosphatase from M. tuberculosis
2381: dUTP pyrophosphatase from M. tuberculosis
Model of an enzyme, dUTP pyrophosphatase, from Mycobacterium tuberculosis. Drugs targeted to this enzyme might inhibit the replication of the bacterium that causes most cases of tuberculosis.
Mycobacterium Tuberculosis Center, PSI
View Media
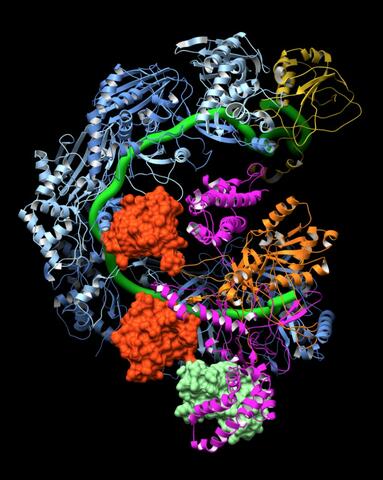
6352: CRISPR surveillance complex
6352: CRISPR surveillance complex
This image shows how the CRISPR surveillance complex is disabled by two copies of anti-CRISPR protein AcrF1 (red) and one AcrF2 (light green). These anti-CRISPRs block access to the CRISPR RNA (green tube) preventing the surveillance complex from scanning and targeting invading viral DNA for destruction.
NRAMM National Resource for Automated Molecular Microscopy http://nramm.nysbc.org/nramm-images/ Source: Bridget Carragher
View Media
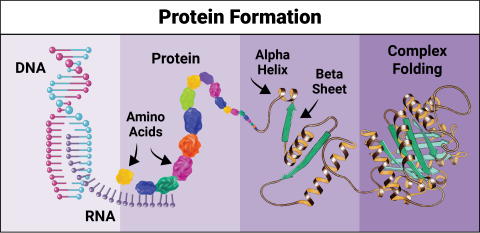
6603: Protein formation
6603: Protein formation
Proteins are 3D structures made up of smaller units. DNA is transcribed to RNA, which in turn is translated into amino acids. Amino acids form a protein strand, which has sections of corkscrew-like coils, called alpha helices, and other sections that fold flat, called beta sheets. The protein then goes through complex folding to produce the 3D structure.
NIGMS, with the folded protein illustration adapted from Jane Richardson, Duke University Medical Center
View Media
2324: Movements of myosin
2324: Movements of myosin
Inside the fertilized egg cell of a fruit fly, we see a type of myosin (related to the protein that helps muscles contract) made to glow by attaching a fluorescent protein. After fertilization, the myosin proteins are distributed relatively evenly near the surface of the embryo. The proteins temporarily vanish each time the cells' nuclei--initially buried deep in the cytoplasm--divide. When the multiplying nuclei move to the surface, they shift the myosin, producing darkened holes. The glowing myosin proteins then gather, contract, and start separating the nuclei into their own compartments.
Victoria Foe, University of Washington
View Media
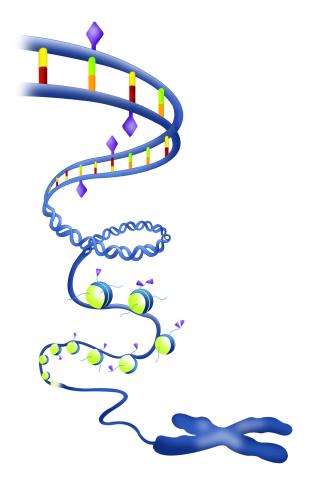
2562: Epigenetic code
2562: Epigenetic code
The "epigenetic code" controls gene activity with chemical tags that mark DNA (purple diamonds) and the "tails" of histone proteins (purple triangles). These markings help determine whether genes will be transcribed by RNA polymerase. Genes hidden from access to RNA polymerase are not expressed. See image 2563 for a labeled version of this illustration. Featured in The New Genetics.
Crabtree + Company
View Media
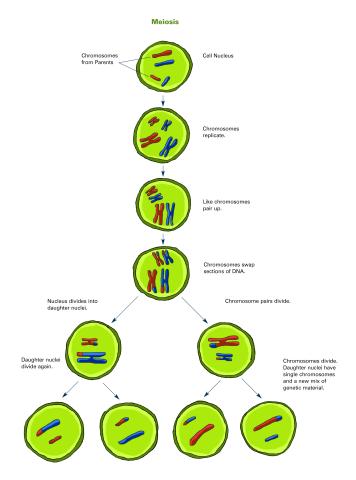
2546: Meiosis illustration (with labels)
2546: Meiosis illustration (with labels)
Meiosis is the process whereby a cell reduces its chromosomes from diploid to haploid in creating eggs or sperm. See image 2545 for an unlabeled version of this illustration. See image 2544 for an unlabeled version of this illustration. Featured in The New Genetics.
Crabtree + Company
View Media
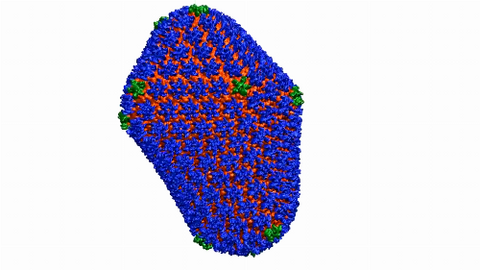
6601: Atomic-level structure of the HIV capsid
6601: Atomic-level structure of the HIV capsid
This animation shows atoms of the HIV capsid, the shell that encloses the virus's genetic material. Scientists determined the exact structure of the capsid using a variety of imaging techniques and analyses. They then entered this data into a supercomputer to produce this image. Related to image 3477.
Juan R. Perilla and the Theoretical and Computational Biophysics Group, University of Illinois at Urbana-Champaign
View Media
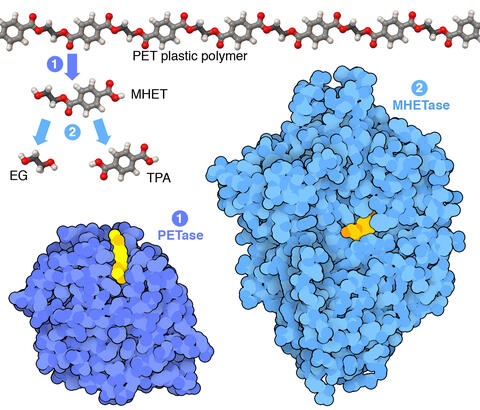
7000: Plastic-eating enzymes
7000: Plastic-eating enzymes
PETase enzyme degrades polyester plastic (polyethylene terephthalate, or PET) into monohydroxyethyl terephthalate (MHET). Then, MHETase enzyme degrades MHET into its constituents ethylene glycol (EG) and terephthalic acid (TPA).
Find these in the RCSB Protein Data Bank: PET hydrolase (PDB entry 5XH3) and MHETase (PDB entry 6QGA).
Find these in the RCSB Protein Data Bank: PET hydrolase (PDB entry 5XH3) and MHETase (PDB entry 6QGA).
Amy Wu and Christine Zardecki, RCSB Protein Data Bank.
View Media
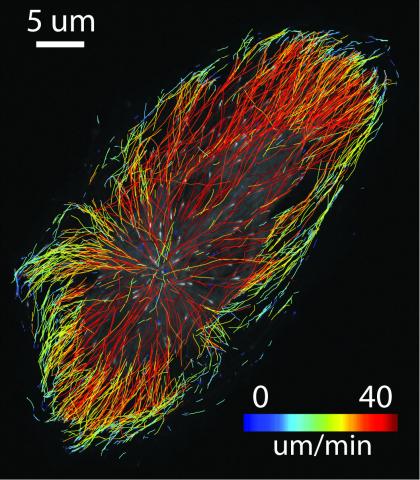
6775: Tracking embryonic zebrafish cells
6775: Tracking embryonic zebrafish cells
To better understand cell movements in developing embryos, researchers isolated cells from early zebrafish embryos and grew them as clusters. Provided with the right signals, the clusters replicated some cell movements seen in intact embryos. Each line in this image depicts the movement of a single cell. The image was created using time-lapse confocal microscopy. Related to video 6776.
Liliana Solnica-Krezel, Washington University School of Medicine in St. Louis.
View Media

1275: Golgi
1275: Golgi
The Golgi complex, also called the Golgi apparatus or, simply, the Golgi. This organelle receives newly made proteins and lipids from the ER, puts the finishing touches on them, addresses them, and sends them to their final destinations.
Judith Stoffer
View Media
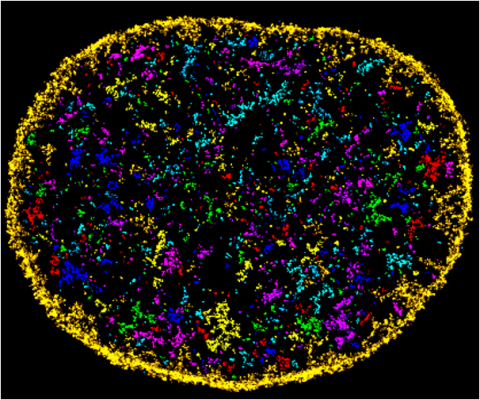
6888: Chromatin in human fibroblast
6888: Chromatin in human fibroblast
The nucleus of a human fibroblast cell with chromatin—a substance made up of DNA and proteins—shown in various colors. Fibroblasts are one of the most common types of cells in mammalian connective tissue, and they play a key role in wound healing and tissue repair. This image was captured using Stochastic Optical Reconstruction Microscopy (STORM).
Related to images 6887 and 6893.
Related to images 6887 and 6893.
Melike Lakadamyali, Perelman School of Medicine at the University of Pennsylvania.
View Media
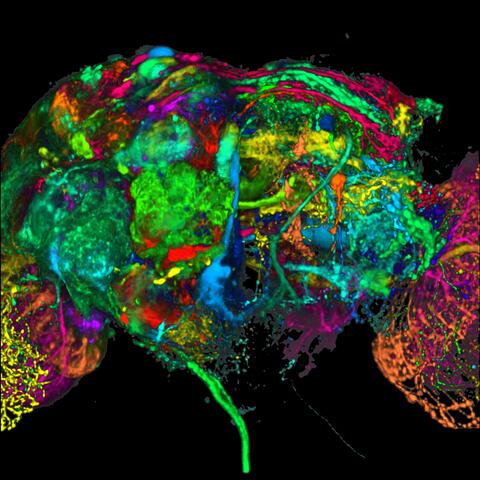
5868: Color coding of the Drosophila brain - black background
5868: Color coding of the Drosophila brain - black background
This image results from a research project to visualize which regions of the adult fruit fly (Drosophila) brain derive from each neural stem cell. First, researchers collected several thousand fruit fly larvae and fluorescently stained a random stem cell in the brain of each. The idea was to create a population of larvae in which each of the 100 or so neural stem cells was labeled at least once. When the larvae grew to adults, the researchers examined the flies’ brains using confocal microscopy. With this technique, the part of a fly’s brain that derived from a single, labeled stem cell “lights up.” The scientists photographed each brain and digitally colorized its lit-up area. By combining thousands of such photos, they created a three-dimensional, color-coded map that shows which part of the Drosophila brain comes from each of its ~100 neural stem cells. In other words, each colored region shows which neurons are the progeny or “clones” of a single stem cell. This work established a hierarchical structure as well as nomenclature for the neurons in the Drosophila brain. Further research will relate functions to structures of the brain.
Related to image 5838 and video 5843.
Related to image 5838 and video 5843.
Yong Wan from Charles Hansen’s lab, University of Utah. Data preparation and visualization by Masayoshi Ito in the lab of Kei Ito, University of Tokyo.
View Media
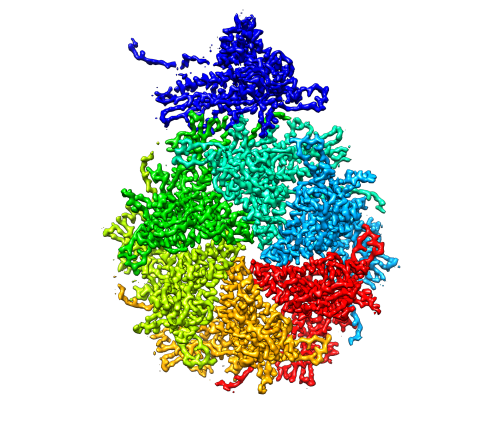
5875: Bacteriophage P22 capsid, detail
5875: Bacteriophage P22 capsid, detail
Detail of a subunit of the capsid, or outer cover, of bacteriophage P22, a virus that infects the Salmonella bacteria. Cryo-electron microscopy (cryo-EM) was used to capture details of the capsid proteins, each shown here in a separate color. Thousands of cryo-EM scans capture the structure and shape of all the individual proteins in the capsid and their position relative to other proteins. A computer model combines these scans into the image shown here. Related to image 5874.
Dr. Wah Chiu, Baylor College of Medicine
View Media
2793: Anti-tumor drug ecteinascidin 743 (ET-743) with hydrogens 04
2793: Anti-tumor drug ecteinascidin 743 (ET-743) with hydrogens 04
Ecteinascidin 743 (ET-743, brand name Yondelis), was discovered and isolated from a sea squirt, Ecteinascidia turbinata, by NIGMS grantee Kenneth Rinehart at the University of Illinois. It was synthesized by NIGMS grantees E.J. Corey and later by Samuel Danishefsky. Multiple versions of this structure are available as entries 2790-2797.
Timothy Jamison, Massachusetts Institute of Technology
View Media
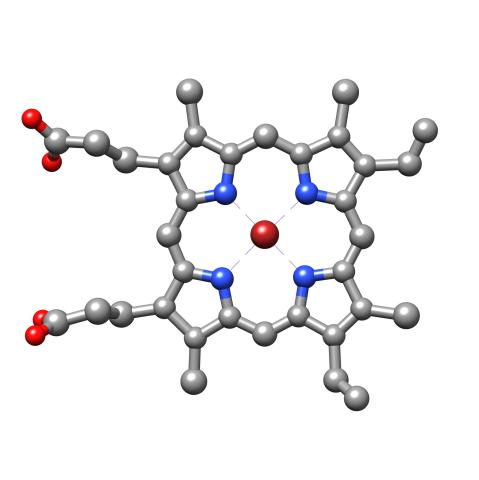
3539: Structure of heme, top view
3539: Structure of heme, top view
Molecular model of the struture of heme. Heme is a small, flat molecule with an iron ion (dark red) at its center. Heme is an essential component of hemoglobin, the protein in blood that carries oxygen throughout our bodies. This image first appeared in the September 2013 issue of Findings Magazine. View side view of heme here 3540.
Rachel Kramer Green, RCSB Protein Data Bank
View Media

1089: Natcher Building 09
1089: Natcher Building 09
NIGMS staff are located in the Natcher Building on the NIH campus.
Alisa Machalek, National Institute of General Medical Sciences
View Media
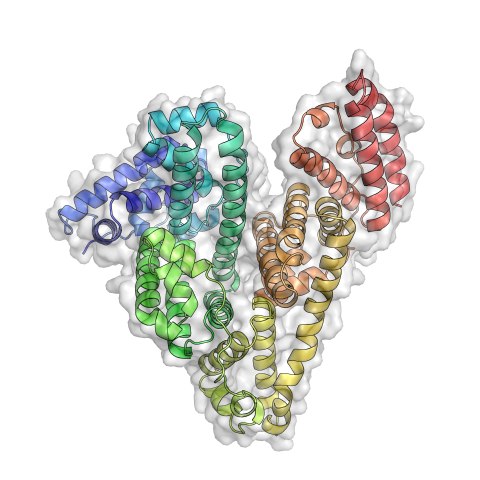
3746: Serum albumin structure 3
3746: Serum albumin structure 3
Serum albumin (SA) is the most abundant protein in the blood plasma of mammals. SA has a characteristic heart-shape structure and is a highly versatile protein. It helps maintain normal water levels in our tissues and carries almost half of all calcium ions in human blood. SA also transports some hormones, nutrients and metals throughout the bloodstream. Despite being very similar to our own SA, those from other animals can cause some mild allergies in people. Therefore, some scientists study SAs from humans and other mammals to learn more about what subtle structural or other differences cause immune responses in the body.
Related to entries 3744 and 3745.
Related to entries 3744 and 3745.
Wladek Minor, University of Virginia
View Media
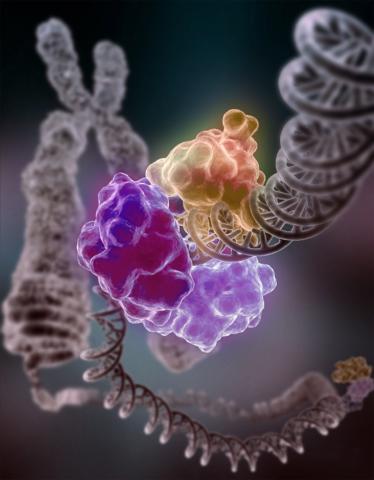
3493: Repairing DNA
3493: Repairing DNA
Like a watch wrapped around a wrist, a special enzyme encircles the double helix to repair a broken strand of DNA. Without molecules that can mend such breaks, cells can malfunction, die, or become cancerous. Related to image 2330.
Tom Ellenberger, Washington University School of Medicine
View Media

3732: A molecular interaction network in yeast 2
3732: A molecular interaction network in yeast 2
The image visualizes a part of the yeast molecular interaction network. The lines in the network represent connections among genes (shown as little dots) and different-colored networks indicate subnetworks, for instance, those in specific locations or pathways in the cell. Researchers use gene or protein expression data to build these networks; the network shown here was visualized with a program called Cytoscape. By following changes in the architectures of these networks in response to altered environmental conditions, scientists can home in on those genes that become central "hubs" (highly connected genes), for example, when a cell encounters stress. They can then further investigate the precise role of these genes to uncover how a cell's molecular machinery deals with stress or other factors. Related to images 3730 and 3733.
Keiichiro Ono, UCSD
View Media
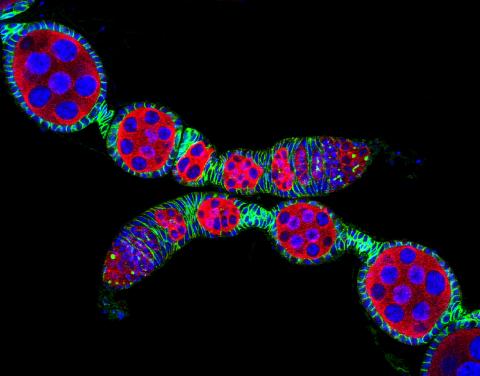
5772: Confocal microscopy image of two Drosophila ovarioles
5772: Confocal microscopy image of two Drosophila ovarioles
Ovarioles in female insects are tubes in which egg cells (called oocytes) form at one end and complete their development as they reach the other end of the tube. This image, taken with a confocal microscope, shows ovarioles in a very popular lab animal, the fruit fly Drosophila. The basic structure of ovarioles supports very rapid egg production, with some insects (like termites) producing several thousand eggs per day. Each insect ovary typically contains four to eight ovarioles, but this number varies widely depending on the insect species.
Scientists use insect ovarioles, for example, to study the basic processes that help various insects, including those that cause disease (like some mosquitos and biting flies), reproduce very quickly.
Scientists use insect ovarioles, for example, to study the basic processes that help various insects, including those that cause disease (like some mosquitos and biting flies), reproduce very quickly.
2004 Olympus BioScapes Competition
View Media
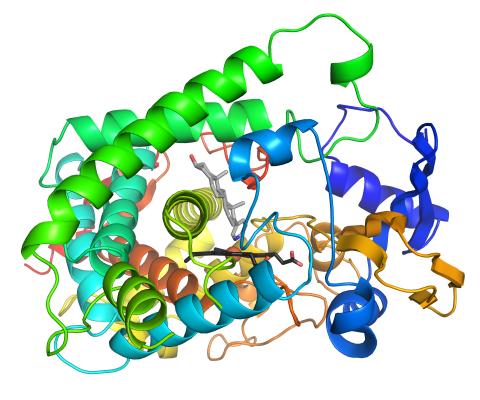
3326: Cytochrome structure with anticancer drug
3326: Cytochrome structure with anticancer drug
This image shows the structure of the CYP17A1 enzyme (ribbons colored from blue N-terminus to red C-terminus), with the associated heme colored black. The prostate cancer drug abiraterone is colored gray. Cytochrome P450 enzymes bind to and metabolize a variety of chemicals, including drugs. Cytochrome P450 17A1 also helps create steroid hormones. Emily Scott's lab is studying how CYP17A1 could be selectively inhibited to treat prostate cancer. She and graduate student Natasha DeVore elucidated the structure shown using X-ray crystallography. Dr. Scott created the image (both white bg and transparent bg) for the NIGMS image gallery. See the "Medium-Resolution Image" for a PNG version of the image that is transparent.
Emily Scott, University of Kansas
View Media
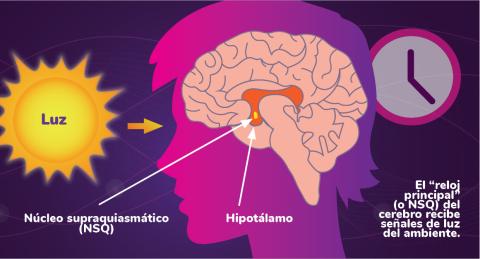
6614: Los ritmos circadianos y el núcleo supraquiasmático
6614: Los ritmos circadianos y el núcleo supraquiasmático
Los ritmos circadianos son cambios físicos, mentales y de comportamiento que siguen un ciclo de 24 horas. Los ritmos circadianos se ven influenciados por la luz y están regulados por el núcleo supraquiasmático del cerebro, a veces denominado el reloj principal.
Vea 6613 para la versión en inglés de esta infografía.
Vea 6613 para la versión en inglés de esta infografía.
NIGMS
View Media
2450: Blood clots show their flex
2450: Blood clots show their flex
Blood clots stop bleeding, but they also can cause heart attacks and strokes. A team led by computational biophysicist Klaus Schulten of the University of Illinois at Urbana-Champaign has revealed how a blood protein can give clots their lifesaving and life-threatening abilities. The researchers combined experimental and computational methods to animate fibrinogen, a protein that forms the elastic fibers that enable clots to withstand the force of blood pressure. This simulation shows that the protein, through a series of events, stretches up to three times its length. Adjusting this elasticity could improve how we manage healthful and harmful clots. NIH's National Center for Research Resources also supported this work. Featured in the March 19, 2008, issue of Biomedical Beat.
Eric Lee, University of Illinois at Urbana-Champaign
View Media
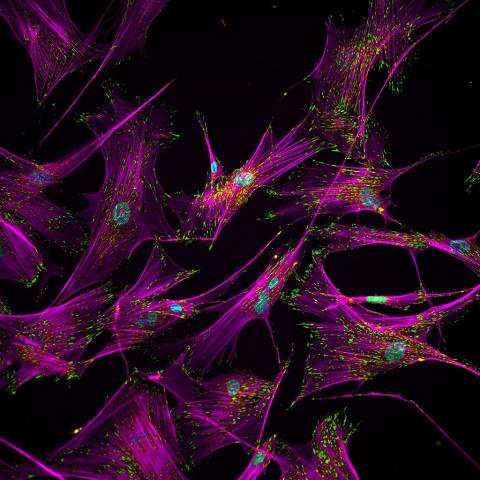
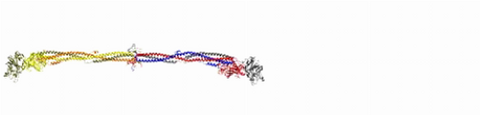
3457: Sticky stem cells
3457: Sticky stem cells
Like a group of barnacles hanging onto a rock, these human cells hang onto a matrix coated glass slide. Actin stress fibers, stained magenta, and the protein vinculin, stained green, make this adhesion possible. The fibroblast nuclei are stained blue.
Ankur Singh and Andrés García, Georgia Institute of Technology
View Media
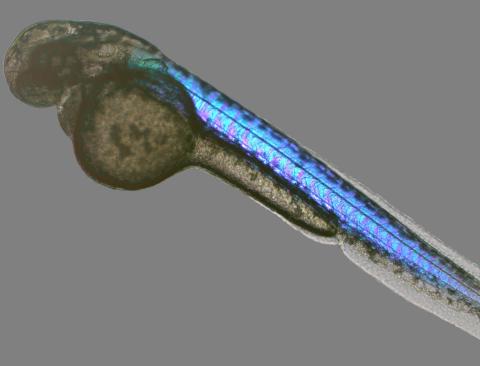
6897: Zebrafish embryo
6897: Zebrafish embryo
A zebrafish embryo showing its natural colors. Zebrafish have see-through eggs and embryos, making them ideal research organisms for studying the earliest stages of development. This image was taken in transmitted light under a polychromatic polarizing microscope.
Michael Shribak, Marine Biological Laboratory/University of Chicago.
View Media

2667: Glowing fish
2667: Glowing fish
Professor Marc Zimmer's family pets, including these fish, glow in the dark in response to blue light. Featured in the September 2009 issue of Findings.
View Media
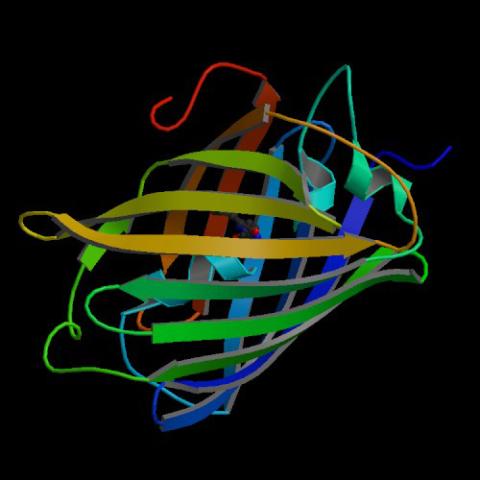
2317: Fruitful dyes
2317: Fruitful dyes
These colorful, computer-generated ribbons show the backbone of a molecule that glows a fluorescent red. The molecule, called mStrawberry, was created by chemists based on a protein found in the ruddy lips of a coral. Scientists use the synthetic molecule and other "fruity" ones like it as a dye to mark and study cell structures.
Roger Y. Tsien, University of California, San Diego
View Media

3530: Lorsch Swearing In
3530: Lorsch Swearing In
Jon Lorsch at his swearing in as NIGMS director in August 2013. Also shown are Francis Collins, NIH Director, and Judith Greenberg, former NIGMS Acting Director.
View Media
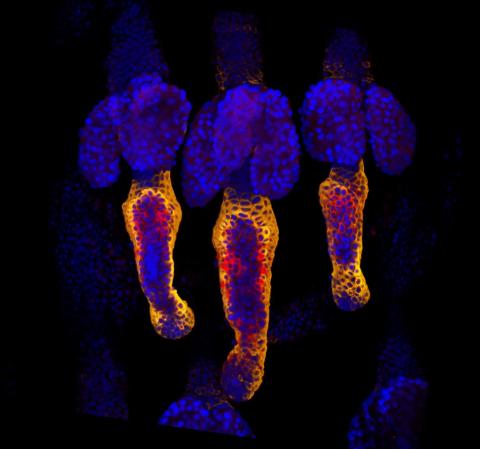
3499: Growing hair follicle stem cells
3499: Growing hair follicle stem cells
Wound healing requires the action of stem cells. In mice that lack the Sept2/ARTS gene, stem cells involved in wound healing live longer and wounds heal faster and more thoroughly than in normal mice. This confocal microscopy image from a mouse lacking the Sept2/ARTS gene shows a tail wound in the process of healing. Cell nuclei are in blue. Red and orange mark hair follicle stem cells (hair follicle stem cells activate to cause hair regrowth, which indicates healing). See more information in the article in Science.
Hermann Steller, Rockefeller University
View Media
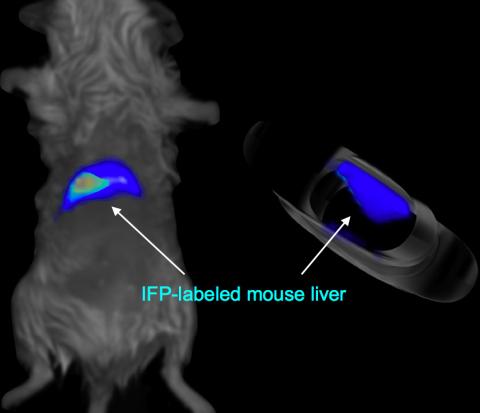
2601: Mouse liver labeled with fluorescent probe
2601: Mouse liver labeled with fluorescent probe
A mouse liver glows after being tagged with specially designed infrared-fluorescent protein (IFP). Since its discovery in 1962, green fluorescent protein (GFP) has become an invaluable resource in biomedical imaging. But because of its short wavelength, the light that makes GFP glow doesn't penetrate far in whole animals. So University of California, San Diego cell biologist Roger Tsien--who shared the 2008 Nobel Prize in chemistry for groundbreaking work with GFP--made infrared-fluorescent proteins (IFPs) that shine under longer-wavelength light, allowing whole-body imaging in small animals.
Xiaokun Shu, University of California, San Diego
View Media
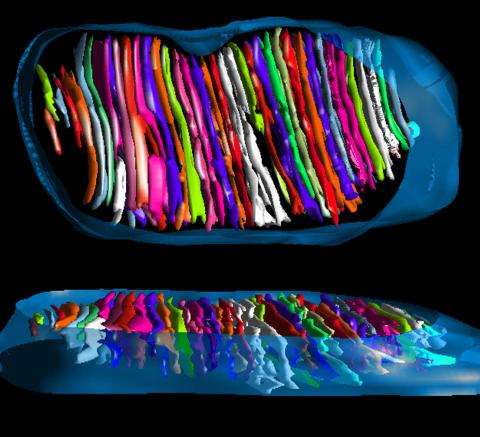
3662: Mitochondrion from insect flight muscle
3662: Mitochondrion from insect flight muscle
This is a tomographic reconstruction of a mitochondrion from an insect flight muscle. Mitochondria are cellular compartments that are best known as the powerhouses that convert energy from the food into energy that runs a range of biological processes. Nearly all our cells have mitochondria.
National Center for Microscopy and Imaging Research
View Media
6518: Biofilm formed by a pathogen
6518: Biofilm formed by a pathogen
A biofilm is a highly organized community of microorganisms that develops naturally on certain surfaces. These communities are common in natural environments and generally do not pose any danger to humans. Many microbes in biofilms have a positive impact on the planet and our societies. Biofilms can be helpful in treatment of wastewater, for example. This dime-sized biofilm, however, was formed by the opportunistic pathogen Pseudomonas aeruginosa. Under some conditions, this bacterium can infect wounds that are caused by severe burns. The bacterial cells release a variety of materials to form an extracellular matrix, which is stained red in this photograph. The matrix holds the biofilm together and protects the bacteria from antibiotics and the immune system.
Scott Chimileski, Ph.D., and Roberto Kolter, Ph.D., Harvard Medical School.
View Media
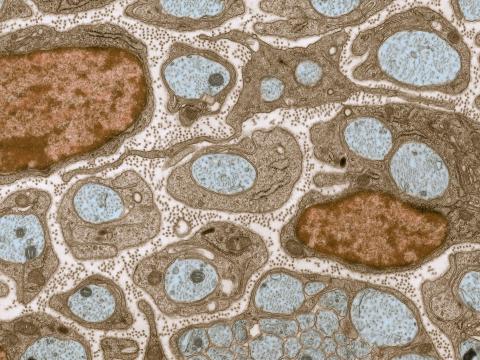
3736: Transmission electron microscopy of myelinated axons with ECM between the axons
3736: Transmission electron microscopy of myelinated axons with ECM between the axons
The extracellular matrix (ECM) is most prevalent in connective tissues but also is present between the stems (axons) of nerve cells, as shown here. Blue-colored nerve cell axons are surrounded by brown-colored, myelin-supplying Schwann cells, which act like insulation around an electrical wire to help speed the transmission of electric nerve impulses down the axon. The ECM is pale pink. The tiny brown spots within it are the collagen fibers that are part of the ECM.
Tom Deerinck, National Center for Microscopy and Imaging Research (NCMIR)
View Media
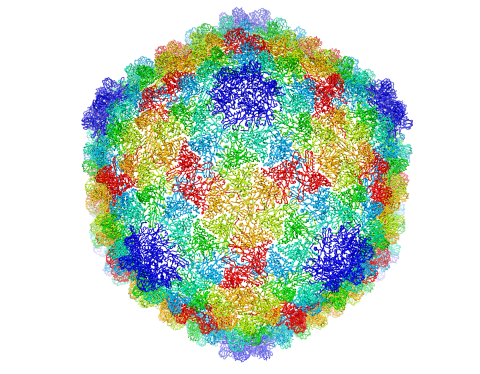
5874: Bacteriophage P22 capsid
5874: Bacteriophage P22 capsid
Cryo-electron microscopy (cryo-EM) has the power to capture details of proteins and other small biological structures at the molecular level. This image shows proteins in the capsid, or outer cover, of bacteriophage P22, a virus that infects the Salmonella bacteria. Each color shows the structure and position of an individual protein in the capsid. Thousands of cryo-EM scans capture the structure and shape of all the individual proteins in the capsid and their position relative to other proteins. A computer model combines these scans into the three-dimension image shown here. Related to image 5875.
Dr. Wah Chiu, Baylor College of Medicine
View Media
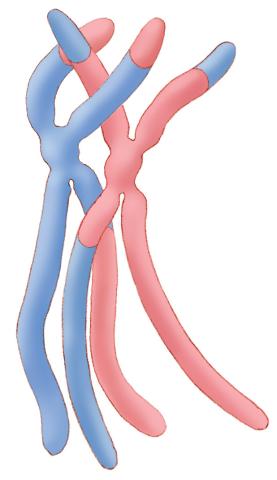
1314: Chromosomes after crossing over
1314: Chromosomes after crossing over
Duplicated pair of chromosomes have exchanged material.
Judith Stoffer
View Media
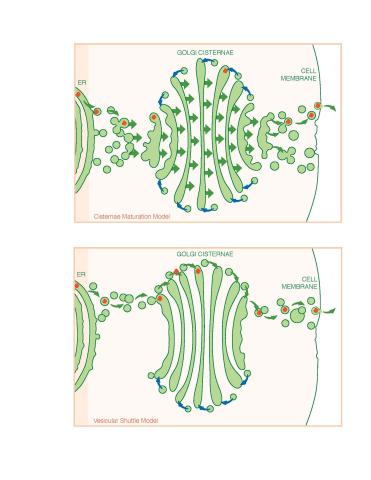
1278: Golgi theories
1278: Golgi theories
Two models for how material passes through the Golgi apparatus: the vesicular shuttle model and the cisternae maturation model.
Judith Stoffer
View Media
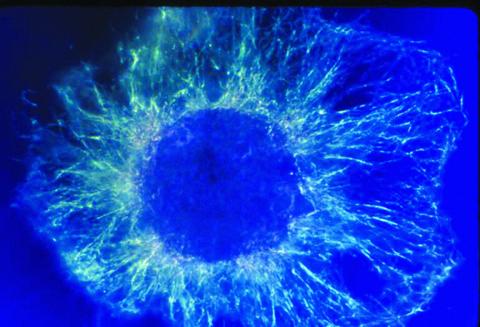
1058: Lily mitosis 01
1058: Lily mitosis 01
A light microscope image shows the chromosomes, stained dark blue, in a dividing cell of an African globe lily (Scadoxus katherinae). This is one frame of a time-lapse sequence that shows cell division in action. The lily is considered a good organism for studying cell division because its chromosomes are much thicker and easier to see than human ones.
Andrew S. Bajer, University of Oregon, Eugene
View Media
6548: Partial Model of a Cilium’s Doublet Microtubule
6548: Partial Model of a Cilium’s Doublet Microtubule
Cilia (cilium in singular) are complex molecular machines found on many of our cells. One component of cilia is the doublet microtubule, a major part of cilia’s skeletons that give them support and shape. This animated image is a partial model of a doublet microtubule’s structure based on cryo-electron microscopy images. Video can be found here 6549.
Brown Lab, Harvard Medical School and Veronica Falconieri Hays.
View Media
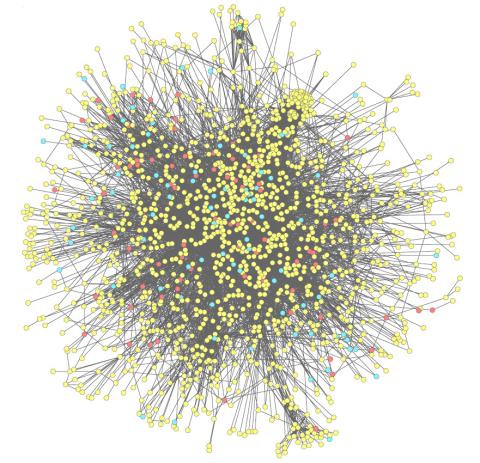
2743: Molecular interactions
2743: Molecular interactions
This network map shows molecular interactions (yellow) associated with a congenital condition that causes heart arrhythmias and the targets for drugs that alter these interactions (red and blue).
Ravi Iyengar, Mount Sinai School of Medicine
View Media
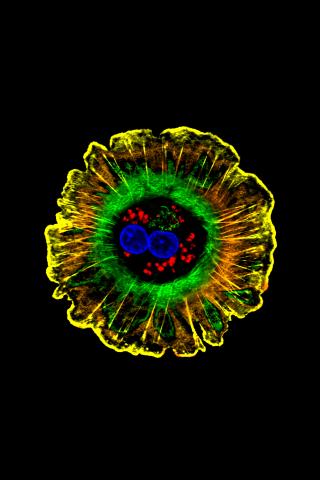
3610: Human liver cell (hepatocyte)
3610: Human liver cell (hepatocyte)
Hepatocytes, like the one shown here, are the most abundant type of cell in the human liver. They play an important role in building proteins; producing bile, a liquid that aids in digesting fats; and chemically processing molecules found normally in the body, like hormones, as well as foreign substances like medicines and alcohol.
This image was part of the Life: Magnified exhibit that ran from June 3, 2014, to January 21, 2015, at Dulles International Airport.
This image was part of the Life: Magnified exhibit that ran from June 3, 2014, to January 21, 2015, at Dulles International Airport.
Donna Beer Stolz, University of Pittsburgh
View Media
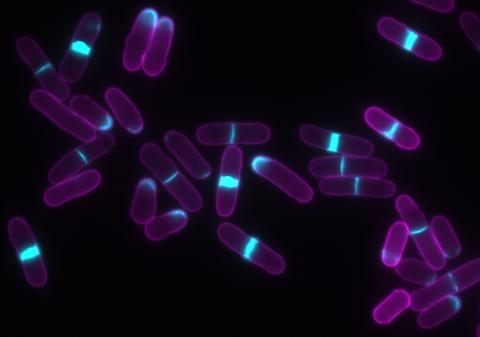
6797: Yeast cells with accumulated cell wall material
6797: Yeast cells with accumulated cell wall material
Yeast cells that abnormally accumulate cell wall material (blue) at their ends and, when preparing to divide, in their middles. This image was captured using wide-field microscopy with deconvolution.
Related to images 6791, 6792, 6793, 6794, 6798, and videos 6795 and 6796.
Related to images 6791, 6792, 6793, 6794, 6798, and videos 6795 and 6796.
Alaina Willet, Kathy Gould’s lab, Vanderbilt University.
View Media

6962: Trigonium diatom
6962: Trigonium diatom
A Trigonium diatom imaged by a quantitative orientation-independent differential interference contrast (OI-DIC) microscope. Diatoms are single-celled photosynthetic algae with mineralized cell walls that contain silica and provide protection and support. These organisms form an important part of the plankton at the base of the marine and freshwater food chains. The width of this image is 90 μm.
More information about the microscopy that produced this image can be found in the Journal of Microscopy paper “An Orientation-Independent DIC Microscope Allows High Resolution Imaging of Epithelial Cell Migration and Wound Healing in a Cnidarian Model” by Malamy and Shribak.
More information about the microscopy that produced this image can be found in the Journal of Microscopy paper “An Orientation-Independent DIC Microscope Allows High Resolution Imaging of Epithelial Cell Migration and Wound Healing in a Cnidarian Model” by Malamy and Shribak.
Michael Shribak, Marine Biological Laboratory/University of Chicago.
View Media
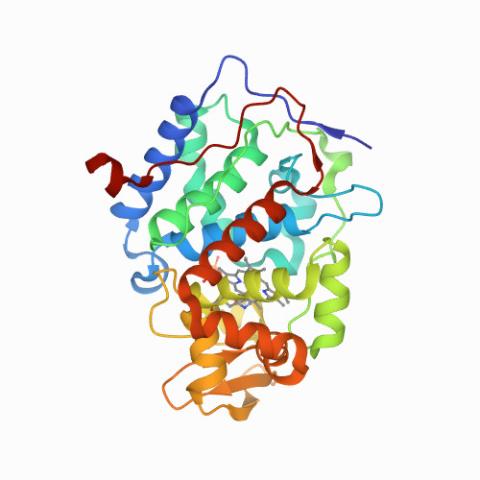
6762: CCP enzyme
6762: CCP enzyme
The enzyme CCP is found in the mitochondria of baker’s yeast. Scientists study the chemical reactions that CCP triggers, which involve a water molecule, iron, and oxygen. This structure was determined using an X-ray free electron laser.
Protein Data Bank.
View Media
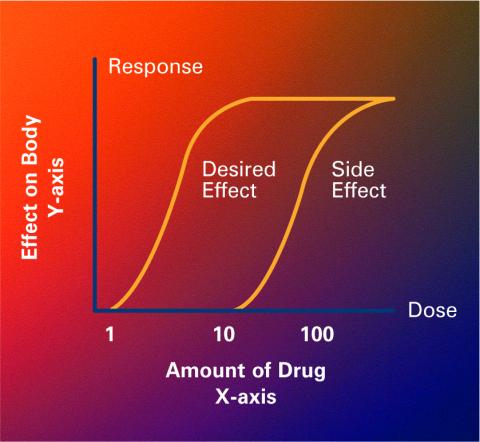
2533: Dose response curves
2533: Dose response curves
Dose-response curves determine how much of a drug (X-axis) causes a particular effect, or a side effect, in the body (Y-axis). Featured in Medicines By Design.
Crabtree + Company
View Media
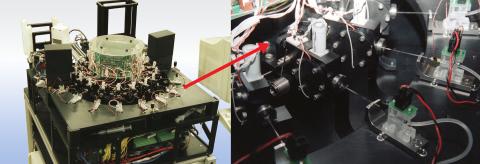
2357: Capillary protein crystallization robot
2357: Capillary protein crystallization robot
This ACAPELLA robot for capillary protein crystallization grows protein crystals, freezes them, and centers them without manual intervention. The close-up is a view of one of the dispensers used for dispensing proteins and reagents.
Structural Genomics of Pathogenic Protozoa Consortium
View Media