Switch to List View
Image and Video Gallery
This is a searchable collection of scientific photos, illustrations, and videos. The images and videos in this gallery are licensed under Creative Commons Attribution Non-Commercial ShareAlike 3.0. This license lets you remix, tweak, and build upon this work non-commercially, as long as you credit and license your new creations under identical terms.
2796: Anti-tumor drug ecteinascidin 743 (ET-743), structure without hydrogens 03
2796: Anti-tumor drug ecteinascidin 743 (ET-743), structure without hydrogens 03
Ecteinascidin 743 (ET-743, brand name Yondelis), was discovered and isolated from a sea squirt, Ecteinascidia turbinata, by NIGMS grantee Kenneth Rinehart at the University of Illinois. It was synthesized by NIGMS grantees E.J. Corey and later by Samuel Danishefsky. Multiple versions of this structure are available as entries 2790-2797.
Timothy Jamison, Massachusetts Institute of Technology
View Media
3449: Calcium uptake during ATP production in mitochondria
3449: Calcium uptake during ATP production in mitochondria
Living primary mouse embryonic fibroblasts. Mitochondria (green) stained with the mitochondrial membrane potential indicator, rhodamine 123. Nuclei (blue) are stained with DAPI. Caption from a November 26, 2012 news release from U Penn (Penn Medicine).
Lili Guo, Perelman School of Medicine, University of Pennsylvania
View Media
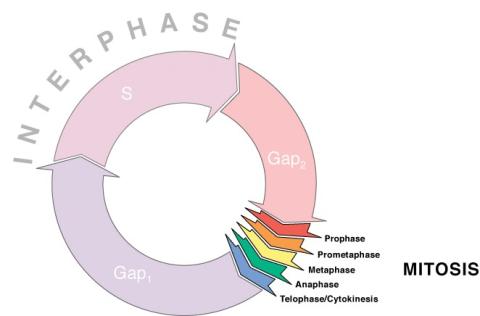
1310: Cell cycle wheel
1310: Cell cycle wheel
A typical animal cell cycle lasts roughly 24 hours, but depending on the type of cell, it can vary in length from less than 8 hours to more than a year. Most of the variability occurs in Gap1. Appears in the NIGMS booklet Inside the Cell.
Judith Stoffer
View Media

2313: Colorful communication
2313: Colorful communication
The marine bacterium Vibrio harveyi glows when near its kind. This luminescence, which results from biochemical reactions, is part of the chemical communication used by the organisms to assess their own population size and distinguish themselves from other types of bacteria. But V. harveyi only light up when part of a large group. This communication, called quorum sensing, speaks for itself here on a lab dish, where more densely packed areas of the bacteria show up blue. Other types of bacteria use quorum sensing to release toxins, trigger disease, and evade the immune system.
Bonnie Bassler, Princeton University
View Media
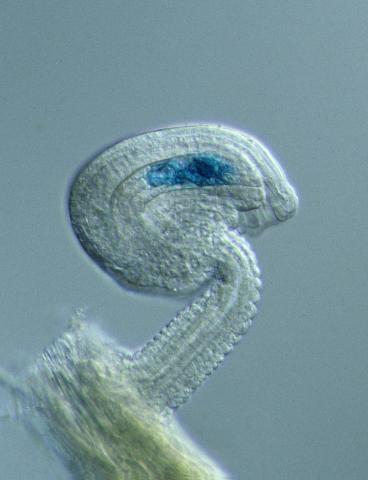
2418: Genetic imprinting in Arabidopsis
2418: Genetic imprinting in Arabidopsis
This delicate, birdlike projection is an immature seed of the Arabidopsis plant. The part in blue shows the cell that gives rise to the endosperm, the tissue that nourishes the embryo. The cell is expressing only the maternal copy of a gene called MEDEA. This phenomenon, in which the activity of a gene can depend on the parent that contributed it, is called genetic imprinting. In Arabidopsis, the maternal copy of MEDEA makes a protein that keeps the paternal copy silent and reduces the size of the endosperm. In flowering plants and mammals, this sort of genetic imprinting is thought to be a way for the mother to protect herself by limiting the resources she gives to any one embryo. Featured in the May 16, 2006, issue of Biomedical Beat.
Robert Fischer, University of California, Berkeley
View Media
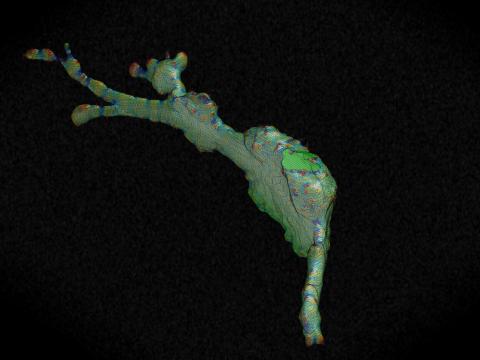
5767: Multivesicular bodies containing intralumenal vesicles assemble at the vacuole 3
5767: Multivesicular bodies containing intralumenal vesicles assemble at the vacuole 3
Collecting and transporting cellular waste and sorting it into recylable and nonrecylable pieces is a complex business in the cell. One key player in that process is the endosome, which helps collect, sort and transport worn-out or leftover proteins with the help of a protein assembly called the endosomal sorting complexes for transport (or ESCRT for short). These complexes help package proteins marked for breakdown into intralumenal vesicles, which, in turn, are enclosed in multivesicular bodies for transport to the places where the proteins are recycled or dumped. In this image, two multivesicular bodies (with yellow membranes) contain tiny intralumenal vesicles (with a diameter of only 25 nanometers; shown in red) adjacent to the cell's vacuole (in orange).
Scientists working with baker's yeast (Saccharomyces cerevisiae) study the budding inward of the limiting membrane (green lines on top of the yellow lines) into the intralumenal vesicles. This tomogram was shot with a Tecnai F-20 high-energy electron microscope, at 29,000x magnification, with a 0.7-nm pixel, ~4-nm resolution.
To learn more about endosomes, see the Biomedical Beat blog post The Cell’s Mailroom. Related to a microscopy photograph 5768 that was used to generate this illustration and a zoomed-out version 5769 of this illustration.
Scientists working with baker's yeast (Saccharomyces cerevisiae) study the budding inward of the limiting membrane (green lines on top of the yellow lines) into the intralumenal vesicles. This tomogram was shot with a Tecnai F-20 high-energy electron microscope, at 29,000x magnification, with a 0.7-nm pixel, ~4-nm resolution.
To learn more about endosomes, see the Biomedical Beat blog post The Cell’s Mailroom. Related to a microscopy photograph 5768 that was used to generate this illustration and a zoomed-out version 5769 of this illustration.
Matthew West and Greg Odorizzi, University of Colorado
View Media
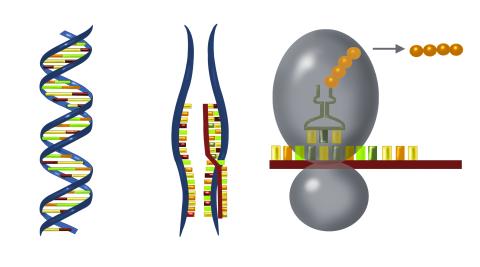
2547: Central dogma, illustrated
2547: Central dogma, illustrated
DNA encodes RNA, which encodes protein. DNA is transcribed to make messenger RNA (mRNA). The mRNA sequence (dark red strand) is complementary to the DNA sequence (blue strand). On ribosomes, transfer RNA (tRNA) reads three nucleotides at a time in mRNA to bring together the amino acids that link up to make a protein. See image 2548 for a labeled version of this illustration and 2549 for a labeled and numbered version. Featured in The New Genetics.
Crabtree + Company
View Media
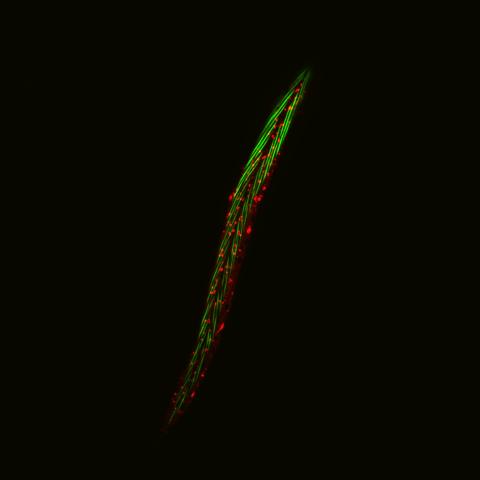
6581: Fluorescent C. elegans showing muscle and ribosomal protein
6581: Fluorescent C. elegans showing muscle and ribosomal protein
C. elegans, a tiny roundworm, with a ribosomal protein glowing red and muscle fibers glowing green. Researchers used these worms to study a molecular pathway that affects aging. The ribosomal protein is involved in protein translation and may play a role in dietary restriction-induced longevity. Image created using confocal microscopy.
View group of roundworms here 6582.
View closeup of roundworms here 6583.
View group of roundworms here 6582.
View closeup of roundworms here 6583.
Jarod Rollins, Mount Desert Island Biological Laboratory.
View Media
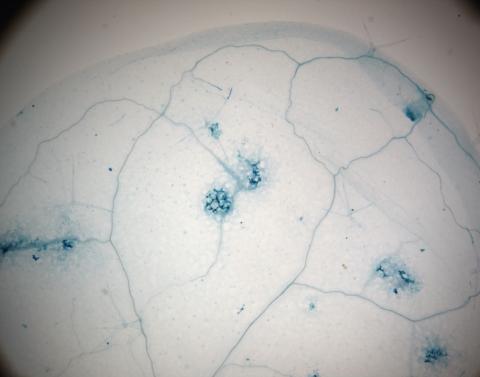
2781: Disease-resistant Arabidopsis leaf
2781: Disease-resistant Arabidopsis leaf
This is a magnified view of an Arabidopsis thaliana leaf a few days after being exposed to the pathogen Hyaloperonospora arabidopsidis. The plant from which this leaf was taken is genetically resistant to the pathogen. The spots in blue show areas of localized cell death where infection occurred, but it did not spread. Compare this response to that shown in Image 2782. Jeff Dangl has been funded by NIGMS to study the interactions between pathogens and hosts that allow or suppress infection.
Jeff Dangl, University of North Carolina, Chapel Hill
View Media
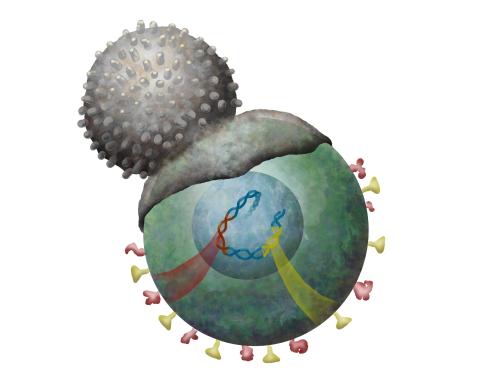
2489: Immune cell attacks cell infected with a retrovirus
2489: Immune cell attacks cell infected with a retrovirus
T cells engulf and digest cells displaying markers (or antigens) for retroviruses, such as HIV.
Kristy Whitehouse, science illustrator
View Media
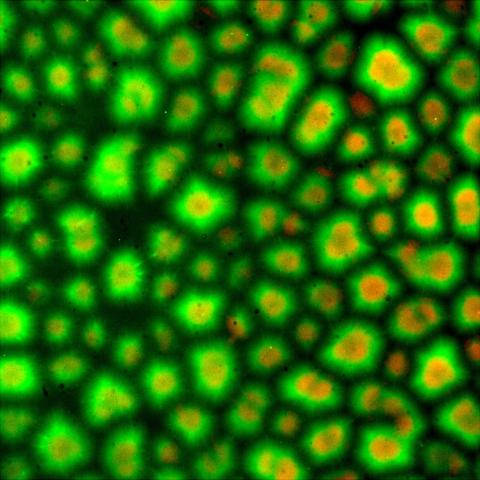
6586: Cell-like compartments from frog eggs 3
6586: Cell-like compartments from frog eggs 3
Cell-like compartments that spontaneously emerged from scrambled frog eggs. Endoplasmic reticulum (red) and microtubules (green) are visible. Image created using epifluorescence microscopy.
For more photos of cell-like compartments from frog eggs view: 6584, 6585, 6591, 6592, and 6593.
For videos of cell-like compartments from frog eggs view: 6587, 6588, 6589, and 6590.
Xianrui Cheng, Stanford University School of Medicine.
View Media
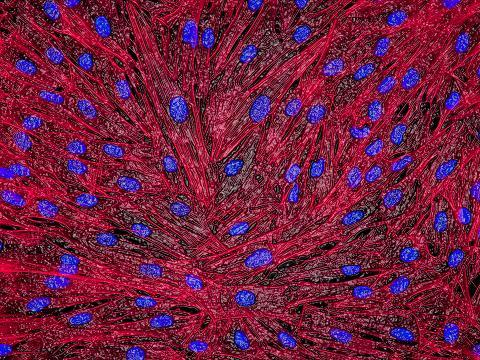
3670: DNA and actin in cultured fibroblast cells
3670: DNA and actin in cultured fibroblast cells
DNA (blue) and actin (red) in cultured fibroblast cells.
Tom Deerinck, National Center for Microscopy and Imaging Research (NCMIR)
View Media

1120: Superconducting magnet
1120: Superconducting magnet
Superconducting magnet for NMR research, from the February 2003 profile of Dorothee Kern in Findings.
Mike Lovett
View Media
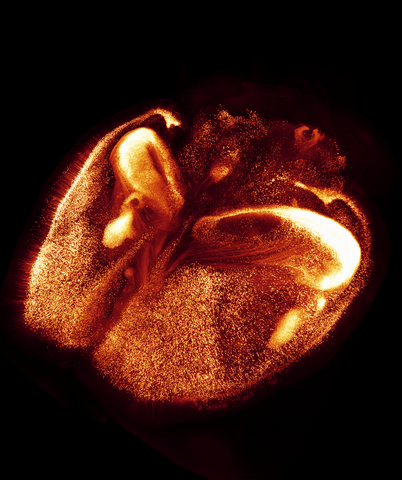
6930: Mouse brain 2
6930: Mouse brain 2
A mouse brain that was genetically modified so that subpopulations of its neurons glow. Researchers often study mice because they share many genes with people and can shed light on biological processes, development, and diseases in humans.
This image was captured using a light sheet microscope.
Related to image 6929 and video 6931.
This image was captured using a light sheet microscope.
Related to image 6929 and video 6931.
Prayag Murawala, MDI Biological Laboratory and Hannover Medical School.
View Media
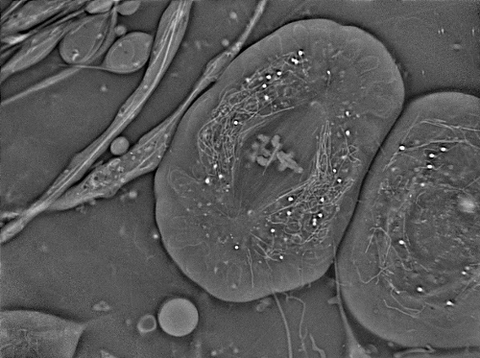
6898: Crane fly spermatocyte undergoing meiosis
6898: Crane fly spermatocyte undergoing meiosis
A crane fly spermatocyte during metaphase of meiosis-I, a step in the production of sperm. A meiotic spindle pulls apart three pairs of autosomal chromosomes, along with a sex chromosome on the right. Tubular mitochondria surround the spindle and chromosomes. This video was captured with quantitative orientation-independent differential interference contrast and is a time lapse showing a 1-second image taken every 30 seconds over the course of 30 minutes.
More information about the research that produced this video can be found in the J. Biomed Opt. paper “Orientation-Independent Differential Interference Contrast (DIC) Microscopy and Its Combination with Orientation-Independent Polarization System” by Shribak et. al.
More information about the research that produced this video can be found in the J. Biomed Opt. paper “Orientation-Independent Differential Interference Contrast (DIC) Microscopy and Its Combination with Orientation-Independent Polarization System” by Shribak et. al.
Michael Shribak, Marine Biological Laboratory/University of Chicago.
View Media
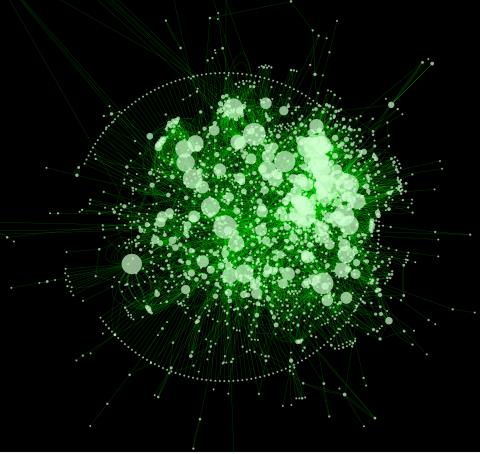
2737: Cytoscape network diagram 1
2737: Cytoscape network diagram 1
Molecular biologists are increasingly relying on bioinformatics software to visualize molecular interaction networks and to integrate these networks with data such as gene expression profiles. Related to 2749.
Keiichiro Ono, Trey Ideker lab, University of California, San Diego
View Media
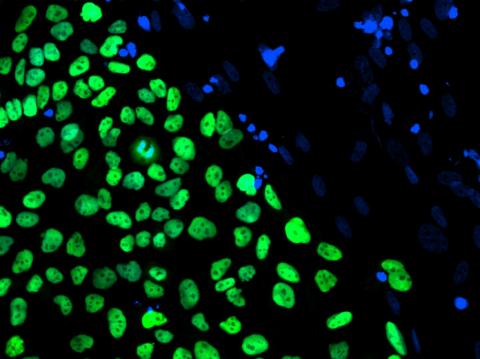
3275: Human embryonic stem cells on feeder cells
3275: Human embryonic stem cells on feeder cells
The nuclei stained green highlight human embryonic stem cells grown under controlled conditions in a laboratory. Blue represents the DNA of surrounding, supportive feeder cells. Image and caption information courtesy of the California Institute for Regenerative Medicine. See related image 3724.
Julie Baker lab, Stanford University School of Medicine, via CIRM
View Media
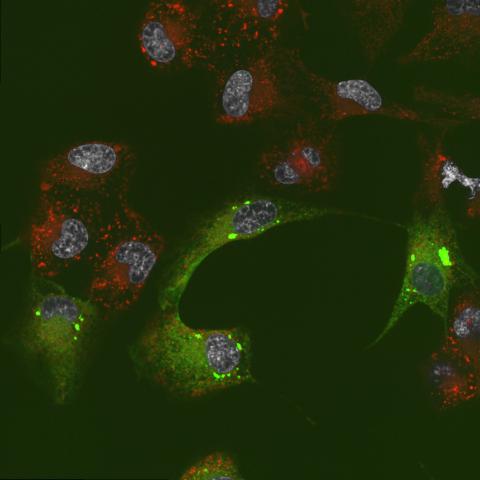
6774: Endoplasmic reticulum abnormalities 2
6774: Endoplasmic reticulum abnormalities 2
Human cells with the gene that codes for the protein FIT2 deleted. After an experimental intervention, they are expressing a nonfunctional version of FIT2, shown in green. The lack of functional FIT2 affected the structure of the endoplasmic reticulum (ER), and the nonfunctional protein clustered in ER membrane aggregates, seen as large bright-green spots. Lipid droplets are shown in red, and the nucleus is visible in gray. This image was captured using a confocal microscope. Related to image 6773.
Michel Becuwe, Harvard University.
View Media
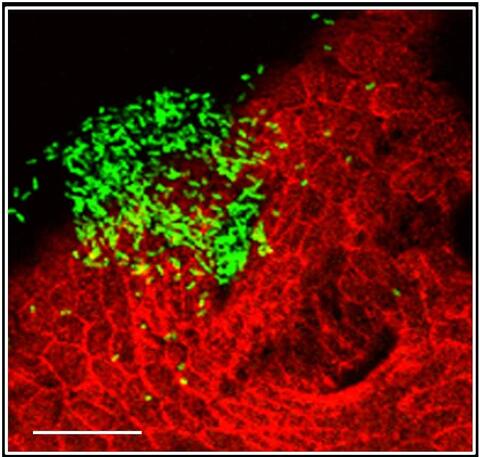
7019: Bacterial cells aggregated above a light-organ pore of the Hawaiian bobtail squid
7019: Bacterial cells aggregated above a light-organ pore of the Hawaiian bobtail squid
The beating of cilia on the outside of the Hawaiian bobtail squid’s light organ concentrates Vibrio fischeri cells (green) present in the seawater into aggregates near the pore-containing tissue (red). From there, the bacterial cells (~2 mm) swim to the pores and migrate through a bottleneck into the interior crypts where a population of symbionts grow and remain for the life of the host. This image was taken using confocal fluorescence microscopy.
Related to images 7016, 7017, 7018, and 7020.
Related to images 7016, 7017, 7018, and 7020.
Margaret J. McFall-Ngai, Carnegie Institution for Science/California Institute of Technology, and Edward G. Ruby, California Institute of Technology.
View Media
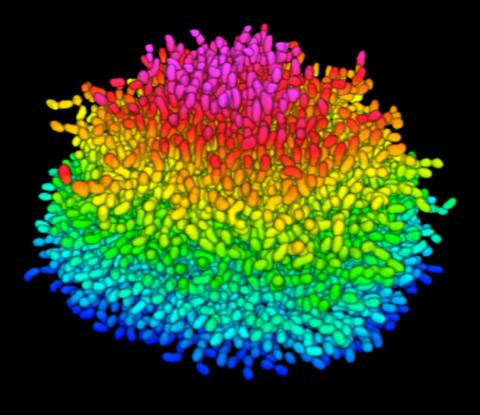
5825: A Growing Bacterial Biofilm
5825: A Growing Bacterial Biofilm
A growing Vibrio cholerae (cholera) biofilm. Cholera bacteria form colonies called biofilms that enable them to resist antibiotic therapy within the body and other challenges to their growth.
Each slightly curved comma shape represents an individual bacterium from assembled confocal microscopy images. Different colors show each bacterium’s position in the biofilm in relation to the surface on which the film is growing.
Each slightly curved comma shape represents an individual bacterium from assembled confocal microscopy images. Different colors show each bacterium’s position in the biofilm in relation to the surface on which the film is growing.
Jing Yan, Ph.D., and Bonnie Bassler, Ph.D., Department of Molecular Biology, Princeton University, Princeton, NJ.
View Media
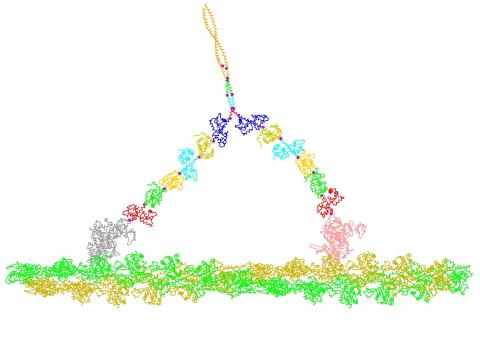
2754: Myosin V binding to actin
2754: Myosin V binding to actin
This simulation of myosin V binding to actin was created using the software tool Protein Mechanica. With Protein Mechanica, researchers can construct models using information from a variety of sources: crystallography, cryo-EM, secondary structure descriptions, as well as user-defined solid shapes, such as spheres and cylinders. The goal is to enable experimentalists to quickly and easily simulate how different parts of a molecule interact.
Simbios, NIH Center for Biomedical Computation at Stanford
View Media
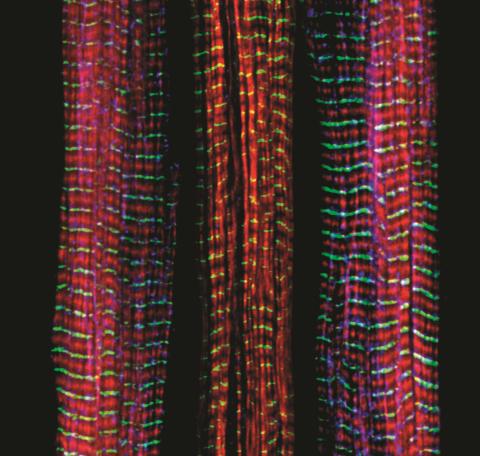
3630: Three muscle fibers; the middle has a defect found in some neuromuscular diseases
3630: Three muscle fibers; the middle has a defect found in some neuromuscular diseases
Of the three muscle fibers shown here, the one on the right and the one on the left are normal. The middle fiber is deficient a large protein called nebulin (blue). Nebulin plays a number of roles in the structure and function of muscles, and its absence is associated with certain neuromuscular disorders.
This image was part of the Life: Magnified exhibit that ran from June 3, 2014, to January 21, 2015, at Dulles International Airport.
This image was part of the Life: Magnified exhibit that ran from June 3, 2014, to January 21, 2015, at Dulles International Airport.
Christopher Pappas and Carol Gregorio, University of Arizona
View Media
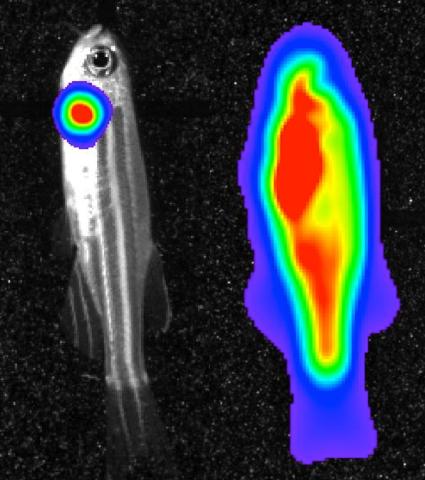
3559: Bioluminescent imaging in adult zebrafish 04
3559: Bioluminescent imaging in adult zebrafish 04
Luciferase-based imaging enables visualization and quantification of internal organs and transplanted cells in live adult zebrafish. This image shows how luciferase-based imaging could be used to visualize the heart for regeneration studies (left), or label all tissues for stem cell transplantation (right).
For imagery of both the lateral and overhead view go to 3556.
For imagery of the overhead view go to 3557.
For imagery of the lateral view go to 3558.
View Media
For imagery of both the lateral and overhead view go to 3556.
For imagery of the overhead view go to 3557.
For imagery of the lateral view go to 3558.
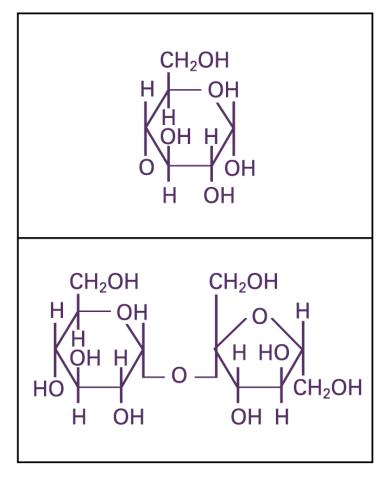
2500: Glucose and sucrose
2500: Glucose and sucrose
Glucose (top) and sucrose (bottom) are sugars made of carbon, hydrogen, and oxygen atoms. Carbohydrates include simple sugars like these and are the main source of energy for the human body.
Crabtree + Company
View Media
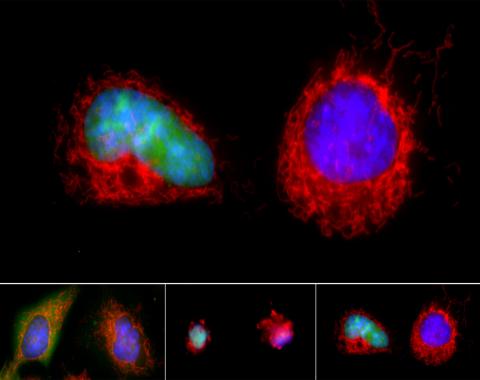
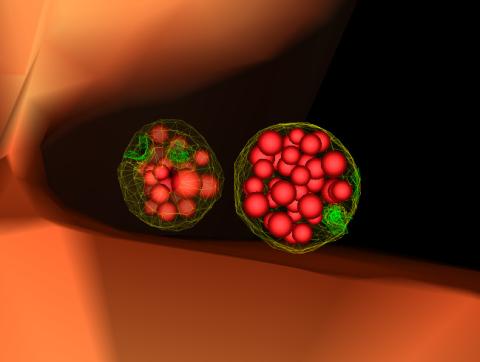
3486: Apoptosis reversed
3486: Apoptosis reversed
Two healthy cells (bottom, left) enter into apoptosis (bottom, center) but spring back to life after a fatal toxin is removed (bottom, right; top).
Hogan Tang of the Denise Montell Lab, Johns Hopkins University School of Medicine
View Media
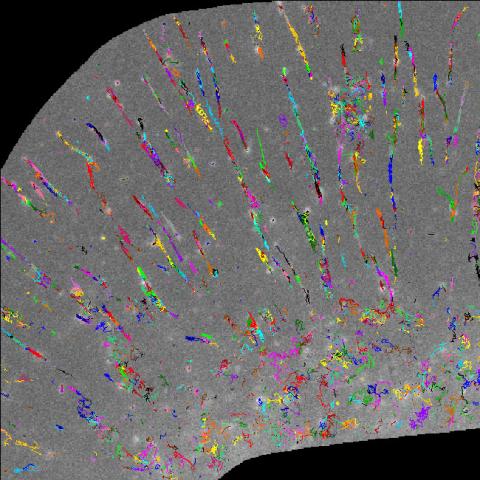
2801: Trajectories of labeled cell receptors
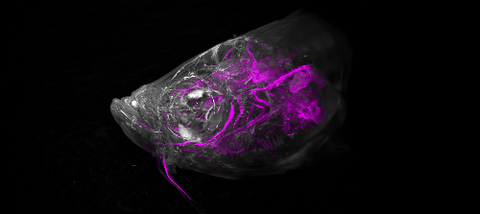
6933: Zebrafish head vasculature video
6933: Zebrafish head vasculature video
Various views of a zebrafish head with blood vessels shown in purple. Researchers often study zebrafish because they share many genes with humans, grow and reproduce quickly, and have see-through eggs and embryos, which make it easy to study early stages of development.
This video was captured using a light sheet microscope.
Related to image 6934.
This video was captured using a light sheet microscope.
Related to image 6934.
Prayag Murawala, MDI Biological Laboratory and Hannover Medical School.
View Media
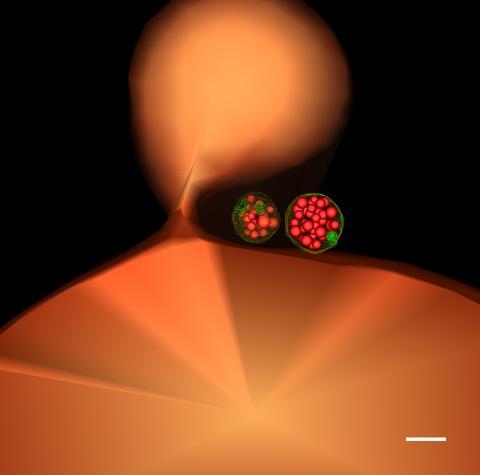
5769: Multivesicular bodies containing intralumenal vesicles assemble at the vacuole 1
5769: Multivesicular bodies containing intralumenal vesicles assemble at the vacuole 1
Collecting and transporting cellular waste and sorting it into recylable and nonrecylable pieces is a complex business in the cell. One key player in that process is the endosome, which helps collect, sort and transport worn-out or leftover proteins with the help of a protein assembly called the endosomal sorting complexes for transport (or ESCRT for short). These complexes help package proteins marked for breakdown into intralumenal vesicles, which, in turn, are enclosed in multivesicular bodies for transport to the places where the proteins are recycled or dumped. In this image, two multivesicular bodies (with yellow membranes) contain tiny intralumenal vesicles (with a diameter of only 25 nanometers; shown in red) adjacent to the cell's vacuole (in orange).
Scientists working with baker's yeast (Saccharomyces cerevisiae) study the budding inward of the limiting membrane (green lines on top of the yellow lines) into the intralumenal vesicles. This tomogram was shot with a Tecnai F-20 high-energy electron microscope, at 29,000x magnification, with a 0.7-nm pixel, ~4-nm resolution.
To learn more about endosomes, see the Biomedical Beat blog post The Cell’s Mailroom. Related to a microscopy photograph 5768 that was used to generate this illustration and a zoomed-in version 5767 of this illustration.
Scientists working with baker's yeast (Saccharomyces cerevisiae) study the budding inward of the limiting membrane (green lines on top of the yellow lines) into the intralumenal vesicles. This tomogram was shot with a Tecnai F-20 high-energy electron microscope, at 29,000x magnification, with a 0.7-nm pixel, ~4-nm resolution.
To learn more about endosomes, see the Biomedical Beat blog post The Cell’s Mailroom. Related to a microscopy photograph 5768 that was used to generate this illustration and a zoomed-in version 5767 of this illustration.
Matthew West and Greg Odorizzi, University of Colorado
View Media
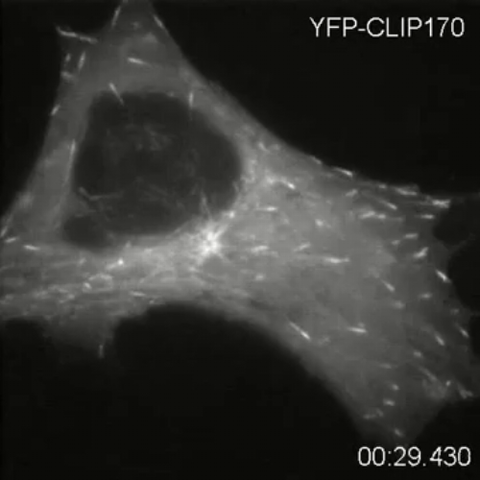
2784: Microtubule dynamics in real time
2784: Microtubule dynamics in real time
Cytoplasmic linker protein (CLIP)-170 is a microtubule plus-end-tracking protein that regulates microtubule dynamics and links microtubule ends to different intracellular structures. In this movie, the gene for CLIP-170 has been fused with green fluorescent protein (GFP). When the protein is expressed in cells, the activities can be monitored in real time. Here, you can see CLIP-170 streaming towards the edges of the cell.
Gary Borisy, Marine Biology Laboratory
View Media
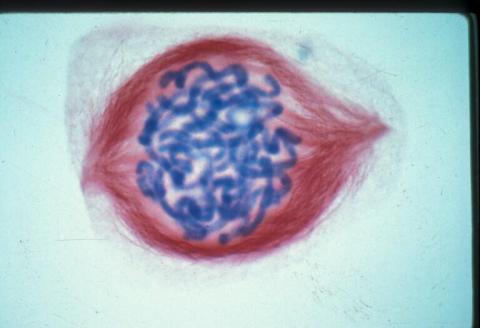
1015: Lily mitosis 05
1015: Lily mitosis 05
A light microscope image of a cell from the endosperm of an African globe lily (Scadoxus katherinae). This is one frame of a time-lapse sequence that shows cell division in action. The lily is considered a good organism for studying cell division because its chromosomes are much thicker and easier to see than human ones. Staining shows microtubules in red and chromosomes in blue. Here, condensed chromosomes are clearly visible.
Related to images 1010, 1011, 1012, 1013, 1014, 1016, 1017, 1018, 1019, and 1021.
Related to images 1010, 1011, 1012, 1013, 1014, 1016, 1017, 1018, 1019, and 1021.
Andrew S. Bajer, University of Oregon, Eugene
View Media
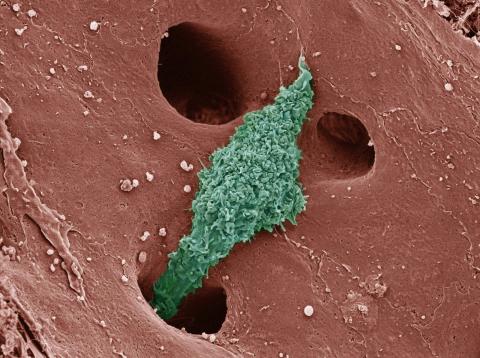
6535: Kupffer cell residing in the liver
6535: Kupffer cell residing in the liver
Kupffer cells appear in the liver during the early stages of mammalian development and stay put throughout life to protect liver cells, clean up old red blood cells, and regulate iron levels. Source article Replenishing the Liver’s Immune Protections. Posted on December 12th, 2019 by Dr. Francis Collins.
Thomas Deerinck, National Center for Microscopy and Imaging Research, University of California, San Diego.
View Media
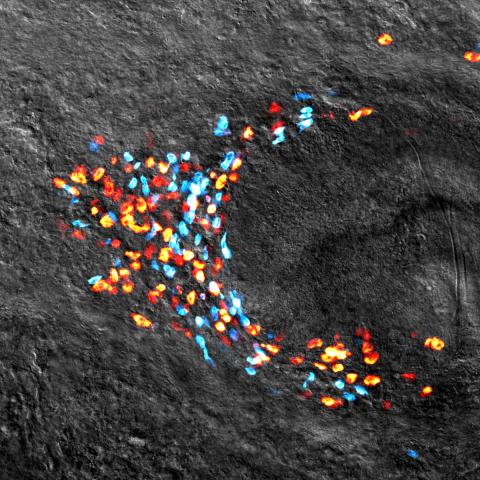
3306: Planarian stem cell colony
3306: Planarian stem cell colony
Planarians are freshwater flatworms that have powerful abilities to regenerate their bodies, which would seem to make them natural model organisms in which to study stem cells. But until recently, scientists had not been able to efficiently find the genes that regulate the planarian stem cell system. In this image, a single stem cell has given rise to a colony of stem cells in a planarian. Proliferating cells are red, and differentiating cells are blue. Quantitatively measuring the size and ratios of these two cell types provides a powerful framework for studying the roles of stem cell regulatory genes in planarians.
Peter Reddien, Whitehead Institute
View Media

7014: Flagellated bacterial cells
7014: Flagellated bacterial cells
Vibrio fischeri (2 mm in length) is the exclusive symbiotic partner of the Hawaiian bobtail squid, Euprymna scolopes. After this bacterium uses its flagella to swim from the seawater into the light organ of a newly hatched juvenile, it colonizes the host and loses the appendages. This image was taken using a scanning electron microscope.
Margaret J. McFall-Ngai, Carnegie Institution for Science/California Institute of Technology, and Edward G. Ruby, California Institute of Technology.
View Media
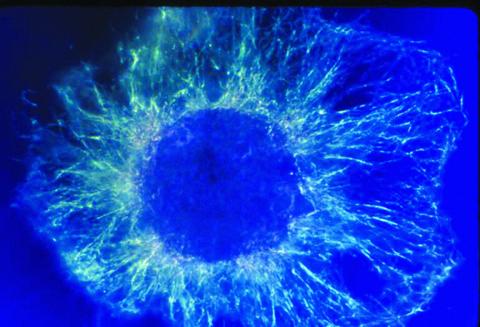
1058: Lily mitosis 01
1058: Lily mitosis 01
A light microscope image shows the chromosomes, stained dark blue, in a dividing cell of an African globe lily (Scadoxus katherinae). This is one frame of a time-lapse sequence that shows cell division in action. The lily is considered a good organism for studying cell division because its chromosomes are much thicker and easier to see than human ones.
Andrew S. Bajer, University of Oregon, Eugene
View Media
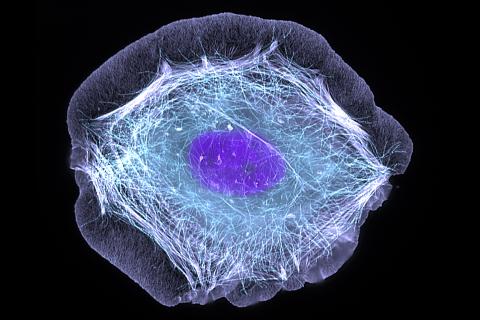
3599: Skin cell (keratinocyte)
3599: Skin cell (keratinocyte)
This normal human skin cell was treated with a growth factor that triggered the formation of specialized protein structures that enable the cell to move. We depend on cell movement for such basic functions as wound healing and launching an immune response.
This image was part of the Life: Magnified exhibit that ran from June 3, 2014, to January 21, 2015, at Dulles International Airport.
This image was part of the Life: Magnified exhibit that ran from June 3, 2014, to January 21, 2015, at Dulles International Airport.
Torsten Wittmann, University of California, San Francisco
View Media
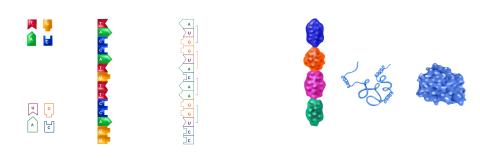
2509: From DNA to Protein
2509: From DNA to Protein
Nucleotides in DNA are copied into RNA, where they are read three at a time to encode the amino acids in a protein. Many parts of a protein fold as the amino acids are strung together.
See image 2510 for a labeled version of this illustration.
Featured in The Structures of Life.
See image 2510 for a labeled version of this illustration.
Featured in The Structures of Life.
Crabtree + Company
View Media
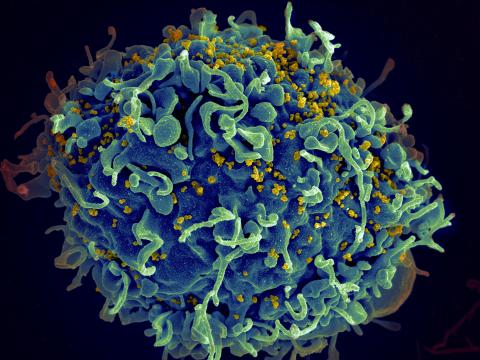
3638: HIV, the AIDS virus, infecting a human cell
3638: HIV, the AIDS virus, infecting a human cell
This human T cell (blue) is under attack by HIV (yellow), the virus that causes AIDS. The virus specifically targets T cells, which play a critical role in the body's immune response against invaders like bacteria and viruses.
This image was part of the Life: Magnified exhibit that ran from June 3, 2014, to January 21, 2015, at Dulles International Airport.
This image was part of the Life: Magnified exhibit that ran from June 3, 2014, to January 21, 2015, at Dulles International Airport.
Seth Pincus, Elizabeth Fischer, and Austin Athman, National Institute of Allergy and Infectious Diseases, National Institutes of Health
View Media
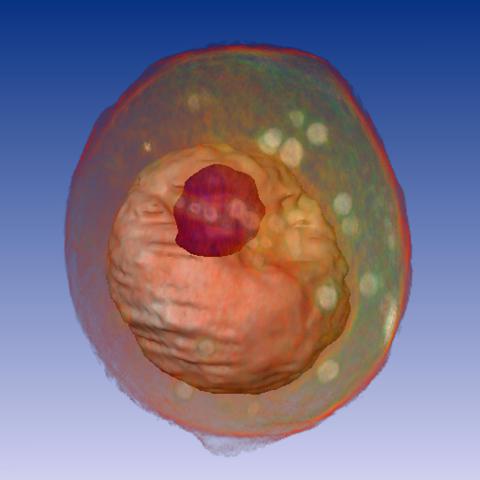
1092: Yeast cell
1092: Yeast cell
A whole yeast (Saccharomyces cerevisiae) cell viewed by X-ray microscopy. Inside, the nucleus and a large vacuole (red) are visible.
Carolyn Larabell, University of California, San Francisco and the Lawrence Berkeley National Laboratory
View Media
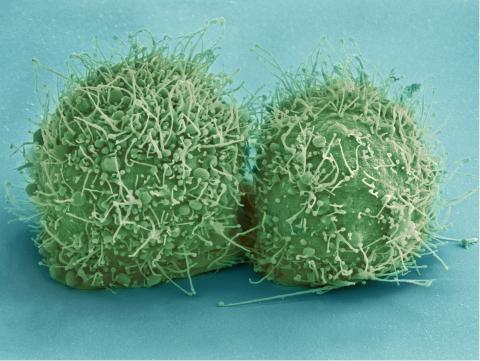
3518: HeLa cells
3518: HeLa cells
Scanning electron micrograph of just-divided HeLa cells. Zeiss Merlin HR-SEM. See related images 3519, 3520, 3521, 3522.
National Center for Microscopy and Imaging Research
View Media
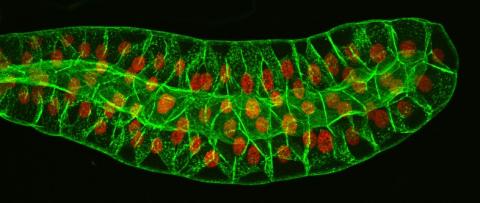
3603: Salivary gland in the developing fruit fly
3603: Salivary gland in the developing fruit fly
For fruit flies, the salivary gland is used to secrete materials for making the pupal case, the protective enclosure in which a larva transforms into an adult fly. For scientists, this gland provided one of the earliest glimpses into the genetic differences between individuals within a species. Chromosomes in the cells of these salivary glands replicate thousands of times without dividing, becoming so huge that scientists can easily view them under a microscope and see differences in genetic content between individuals.
This image was part of the Life: Magnified exhibit that ran from June 3, 2014, to January 21, 2015, at Dulles International Airport.
This image was part of the Life: Magnified exhibit that ran from June 3, 2014, to January 21, 2015, at Dulles International Airport.
Richard Fehon, University of Chicago
View Media
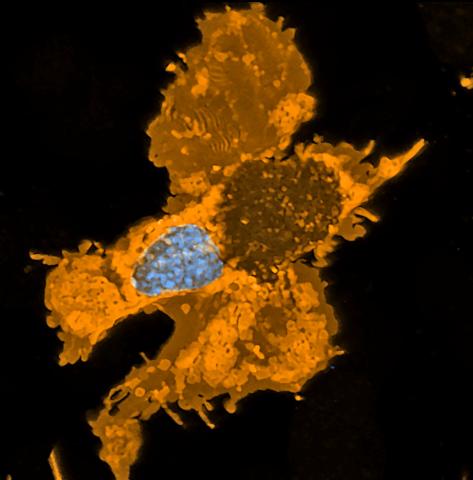
5754: Zebrafish pigment cell
5754: Zebrafish pigment cell
Pigment cells are cells that give skin its color. In fishes and amphibians, like frogs and salamanders, pigment cells are responsible for the characteristic skin patterns that help these organisms to blend into their surroundings or attract mates. The pigment cells are derived from neural crest cells, which are cells originating from the neural tube in the early embryo. Investigating pigment cell formation and migration in animals helps answer important fundamental questions about the factors that control pigmentation in the skin of animals, including humans. This image shows a pigment cell from zebrafish at high resolution. Related to images 5755, 5756, 5757 and 5758.
David Parichy, University of Washington
View Media
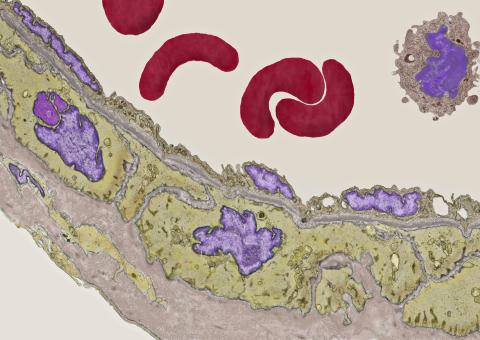
3738: Transmission electron microscopy of coronary artery wall with elastin-rich ECM pseudocolored in light brown
3738: Transmission electron microscopy of coronary artery wall with elastin-rich ECM pseudocolored in light brown
Elastin is a fibrous protein in the extracellular matrix (ECM). It is abundant in artery walls like the one shown here. As its name indicates, elastin confers elasticity. Elastin fibers are at least five times stretchier than rubber bands of the same size. Tissues that expand, such as blood vessels and lungs, need to be both strong and elastic, so they contain both collagen (another ECM protein) and elastin. In this photo, the elastin-rich ECM is colored grayish brown and is most visible at the bottom of the photo. The curved red structures near the top of the image are red blood cells.
Tom Deerinck, National Center for Microscopy and Imaging Research (NCMIR)
View Media
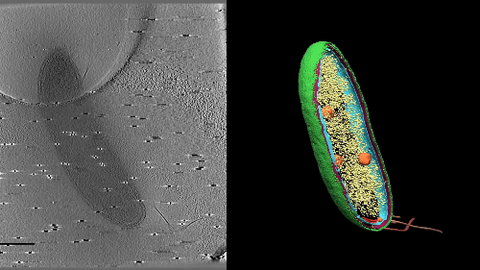
6569: Cryo-electron tomography of a Caulobacter bacterium
6569: Cryo-electron tomography of a Caulobacter bacterium
3D image of Caulobacter bacterium with various components highlighted: cell membranes (red and blue), protein shell (green), protein factories known as ribosomes (yellow), and storage granules (orange).
Peter Dahlberg, Stanford University.
View Media
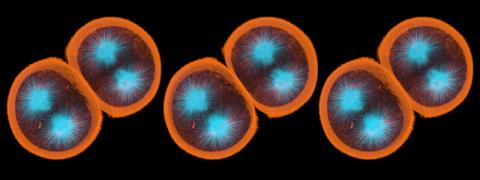
1047: Sea urchin embryo 01
1047: Sea urchin embryo 01
Stereo triplet of a sea urchin embryo stained to reveal actin filaments (orange) and microtubules (blue). This image is part of a series of images: image 1048, image 1049, image 1050, image 1051 and image 1052.
George von Dassow, University of Washington
View Media
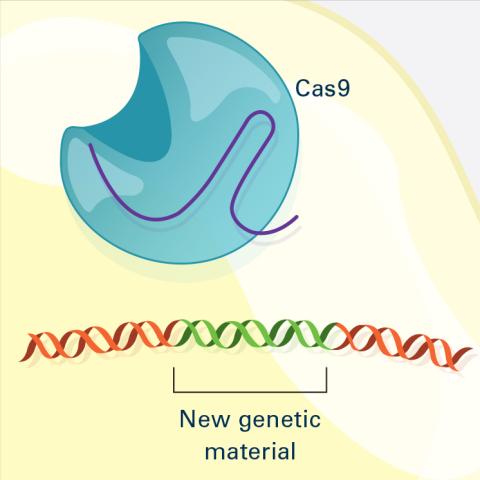
6488: CRISPR Illustration Frame 4
6488: CRISPR Illustration Frame 4
This illustration shows, in simplified terms, how the CRISPR-Cas9 system can be used as a gene-editing tool. The CRISPR system has two components joined together: a finely tuned targeting device (a small strand of RNA programmed to look for a specific DNA sequence) and a strong cutting device (an enzyme called Cas9 that can cut through a double strand of DNA). This frame (4 out of 4) shows a repaired DNA strand with new genetic material that researchers can introduce, which the cell automatically incorporates into the gap when it repairs the broken DNA.
For an explanation and overview of the CRISPR-Cas9 system, see the iBiology video, and find the full CRIPSR illustration here.
For an explanation and overview of the CRISPR-Cas9 system, see the iBiology video, and find the full CRIPSR illustration here.
National Institute of General Medical Sciences.
View Media
3292: Centrioles anchor cilia in planaria
3292: Centrioles anchor cilia in planaria
Centrioles (green) anchor cilia (red), which project on the surface of pharynx cells of the freshwater planarian Schmidtea mediterranea. Centrioles require cellular structures called centrosomes for assembly in other animal species, but this flatworm known for its regenerative ability was unexpectedly found to lack centrosomes. From a Stowers University news release.
Juliette Azimzadeh, University of California, San Francisco
View Media
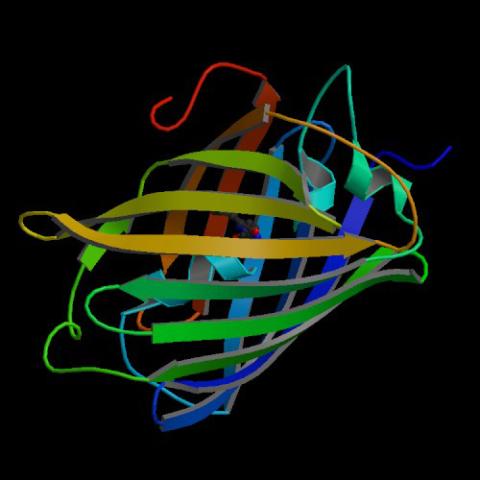
2317: Fruitful dyes
2317: Fruitful dyes
These colorful, computer-generated ribbons show the backbone of a molecule that glows a fluorescent red. The molecule, called mStrawberry, was created by chemists based on a protein found in the ruddy lips of a coral. Scientists use the synthetic molecule and other "fruity" ones like it as a dye to mark and study cell structures.
Roger Y. Tsien, University of California, San Diego
View Media
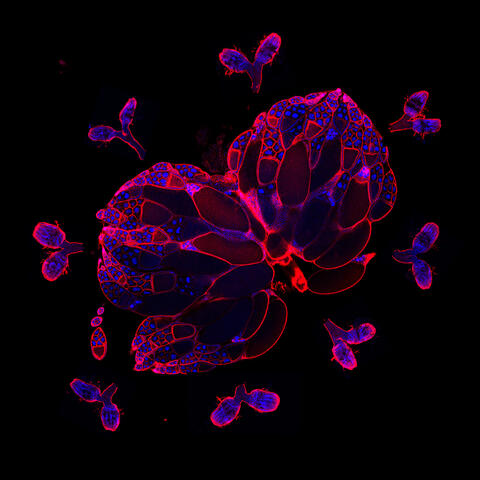
6806: Wild-type and mutant fruit fly ovaries
6806: Wild-type and mutant fruit fly ovaries
The two large, central, round shapes are ovaries from a typical fruit fly (Drosophila melanogaster). The small butterfly-like structures surrounding them are fruit fly ovaries where researchers suppressed the expression of a gene that controls microtubule polymerization and is necessary for normal development. This image was captured using a confocal laser scanning microscope.
Related to image 6807.
Related to image 6807.
Vladimir I. Gelfand, Feinberg School of Medicine, Northwestern University.
View Media
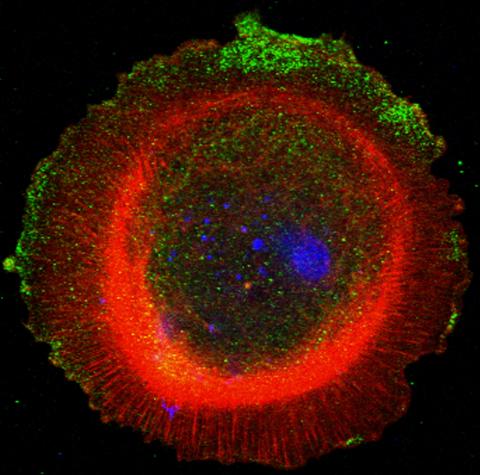
3330: mDia1 antibody staining-01
3330: mDia1 antibody staining-01
Cells move forward with lamellipodia and filopodia supported by networks and bundles of actin filaments. Proper, controlled cell movement is a complex process. Recent research has shown that an actin-polymerizing factor called the Arp2/3 complex is the key component of the actin polymerization engine that drives amoeboid cell motility. ARPC3, a component of the Arp2/3 complex, plays a critical role in actin nucleation. In this photo, the ARPC3+/+ fibroblast cells were fixed and stained with Alexa 546 phalloidin for F-actin (red), mDia1 (green), and DAPI to visualize the nucleus (blue). mDia1 is localized at the lamellipodia of ARPC3+/+ fibroblast cells. Related to images 3328, 3329, 3331, 3332, and 3333.
Rong Li and Praveen Suraneni, Stowers Institute for Medical Research
View Media