Switch to List View
Image and Video Gallery
This is a searchable collection of scientific photos, illustrations, and videos. The images and videos in this gallery are licensed under Creative Commons Attribution Non-Commercial ShareAlike 3.0. This license lets you remix, tweak, and build upon this work non-commercially, as long as you credit and license your new creations under identical terms.
6556: Floral pattern in a mixture of two bacterial species, Acinetobacter baylyi and Escherichia coli, grown on a semi-solid agar for 72 hour
6556: Floral pattern in a mixture of two bacterial species, Acinetobacter baylyi and Escherichia coli, grown on a semi-solid agar for 72 hour
Floral pattern emerging as two bacterial species, motile Acinetobacter baylyi and non-motile Escherichia coli (green), are grown together for 72 hours on 0.5% agar surface from a small inoculum in the center of a Petri dish.
See 6557 for a photo of this process at 24 hours on 0.75% agar surface.
See 6553 for a photo of this process at 48 hours on 1% agar surface.
See 6555 for another photo of this process at 48 hours on 1% agar surface.
See 6550 for a video of this process.
See 6557 for a photo of this process at 24 hours on 0.75% agar surface.
See 6553 for a photo of this process at 48 hours on 1% agar surface.
See 6555 for another photo of this process at 48 hours on 1% agar surface.
See 6550 for a video of this process.
L. Xiong et al, eLife 2020;9: e48885
View Media
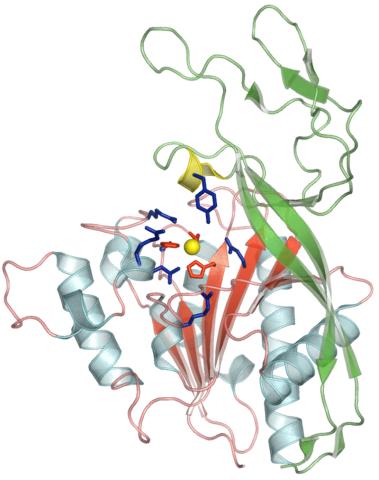
2352: Human aspartoacylase
2352: Human aspartoacylase
Model of aspartoacylase, a human enzyme involved in brain metabolism.
Center for Eukaryotic Structural Genomics, PSI
View Media
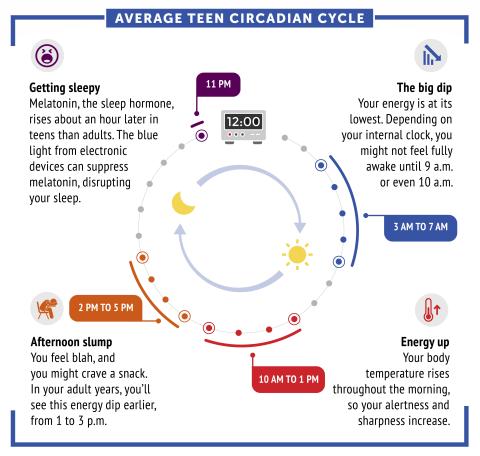
6611: Average teen circadian cycle
6611: Average teen circadian cycle
Circadian rhythms are physical, mental, and behavioral changes that follow a 24-hour cycle. Typical circadian rhythms lead to high energy during the middle of the day (10 a.m. to 1 p.m.) and an afternoon slump. At night, circadian rhythms cause the hormone melatonin to rise, making a person sleepy.
Learn more in NIGMS’ circadian rhythms featured topics page.
See 6612 for the Spanish version of this infographic.
Learn more in NIGMS’ circadian rhythms featured topics page.
See 6612 for the Spanish version of this infographic.
NIGMS
View Media
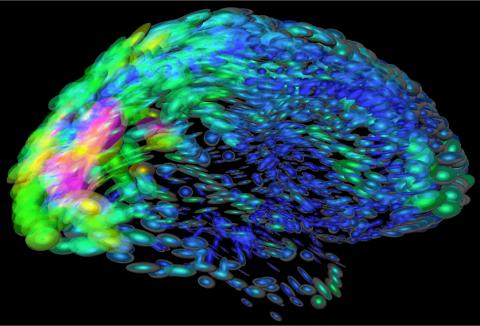
2419: Mapping brain differences
2419: Mapping brain differences
This image of the human brain uses colors and shapes to show neurological differences between two people. The blurred front portion of the brain, associated with complex thought, varies most between the individuals. The blue ovals mark areas of basic function that vary relatively little. Visualizations like this one are part of a project to map complex and dynamic information about the human brain, including genes, enzymes, disease states, and anatomy. The brain maps represent collaborations between neuroscientists and experts in math, statistics, computer science, bioinformatics, imaging, and nanotechnology.
Arthur Toga, University of California, Los Angeles
View Media
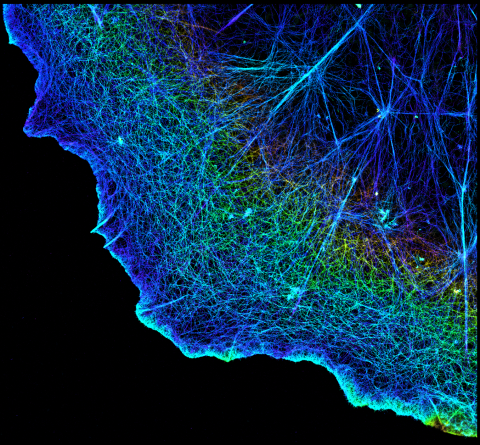
3749: 3D image of actin in a cell
3749: 3D image of actin in a cell
Actin is an essential protein in a cell's skeleton (cytoskeleton). It forms a dense network of thin filaments in the cell. Here, researchers have used a technique called stochastic optical reconstruction microscopy (STORM) to visualize the actin network in a cell in three dimensions. The actin strands were labeled with a dye called Alexa Fluor 647-phalloidin. This image appears in a study published by Nature Methods, which reports how researchers use STORM to visualize the cytoskeleton.
Xiaowei Zhuang, Howard Hughes Medical Institute, Harvard University
View Media
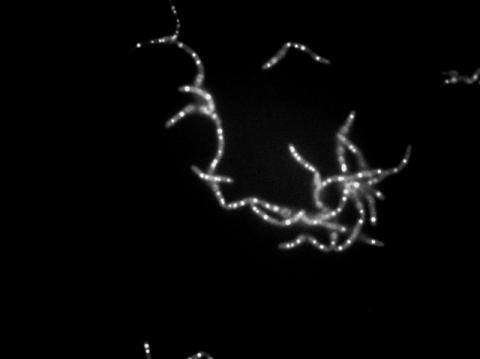
3481: Bacillus anthracis being killed
3481: Bacillus anthracis being killed
Bacillus anthracis (anthrax) cells being killed by a fluorescent trans-translation inhibitor, which disrupts bacterial protein synthesis. The inhibitor is naturally fluorescent and looks blue when it is excited by ultraviolet light in the microscope. This is a black-and-white version of Image 3525.
John Alumasa, Keiler Laboratory, Pennsylvania State University
View Media
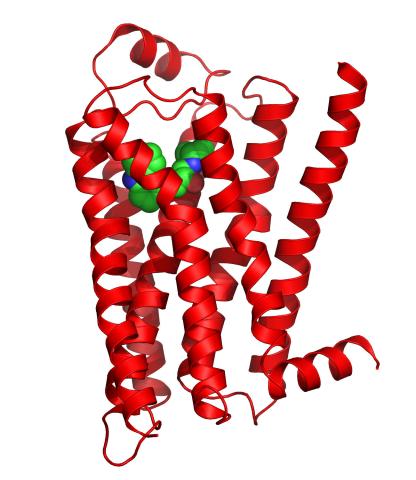
3358: Beta 2-adrenergic receptor
3358: Beta 2-adrenergic receptor
The receptor is shown bound to a partial inverse agonist, carazolol.
Raymond Stevens, The Scripps Research Institute
View Media
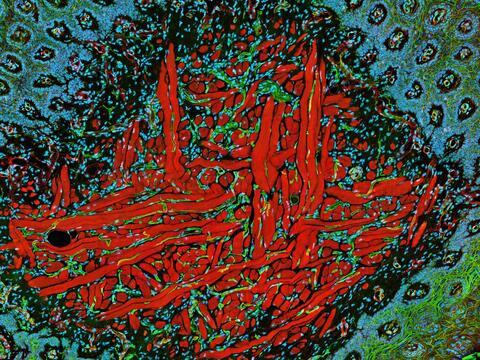
5811: NCMIR Tongue 2
5811: NCMIR Tongue 2
Microscopy image of a tongue. One in a series of two, see image 5810
National Center for Microscopy and Imaging Research (NCMIR)
View Media
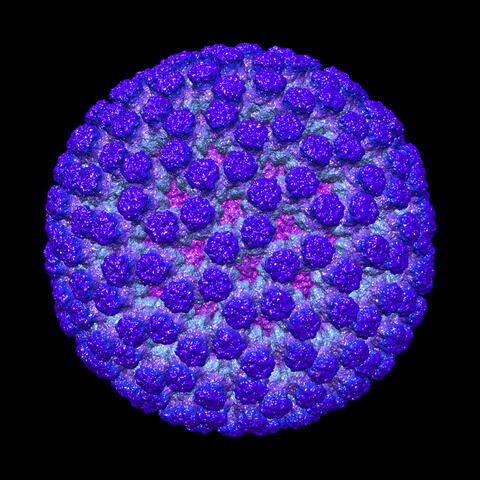
3584: Rotavirus structure
3584: Rotavirus structure
This image shows a computer-generated, three-dimensional map of the rotavirus structure. This virus infects humans and other animals and causes severe diarrhea in infants and young children. By the age of five, almost every child in the world has been infected with this virus at least once. Scientists have found a vaccine against rotavirus, so in the United States there are very few fatalities, but in developing countries and in places where the vaccine is unavailable, this virus is responsible for more than 200,000 deaths each year.
The rotavirus comprises three layers: the outer, middle and inner layers. On infection, the outer layer is removed, leaving behind a "double-layered particle." Researchers have studied the structure of this double-layered particle with a transmission electron microscope. Many images of the virus at a magnification of ~50,000x were acquired, and computational analysis was used to combine the individual particle images into a three-dimensional reconstruction.
The image was rendered by Melody Campbell (PhD student at TSRI). Work that led to the 3D map was published in Campbell et al. Movies of ice-embedded particles enhance resolution in electron cryo-microscopy. Structure. 2012;20(11):1823-8. PMCID: PMC3510009.
This image was part of the Life: Magnified exhibit that ran from June 3, 2014, to January 21, 2015, at Dulles International Airport.
The rotavirus comprises three layers: the outer, middle and inner layers. On infection, the outer layer is removed, leaving behind a "double-layered particle." Researchers have studied the structure of this double-layered particle with a transmission electron microscope. Many images of the virus at a magnification of ~50,000x were acquired, and computational analysis was used to combine the individual particle images into a three-dimensional reconstruction.
The image was rendered by Melody Campbell (PhD student at TSRI). Work that led to the 3D map was published in Campbell et al. Movies of ice-embedded particles enhance resolution in electron cryo-microscopy. Structure. 2012;20(11):1823-8. PMCID: PMC3510009.
This image was part of the Life: Magnified exhibit that ran from June 3, 2014, to January 21, 2015, at Dulles International Airport.
Bridget Carragher, The Scripps Research Institute, La Jolla, CA
View Media
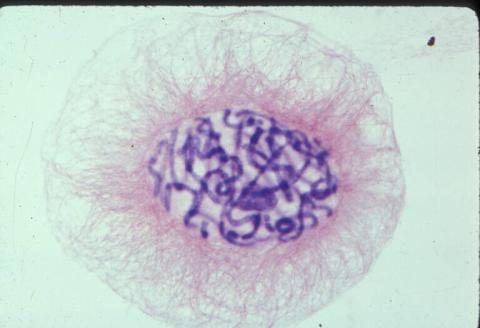
1014: Lily mitosis 04
1014: Lily mitosis 04
A light microscope image of a cell from the endosperm of an African globe lily (Scadoxus katherinae). This is one frame of a time-lapse sequence that shows cell division in action. The lily is considered a good organism for studying cell division because its chromosomes are much thicker and easier to see than human ones. Staining shows microtubules in red and chromosomes in blue.
Related to images 1010, 1011, 1012, 1013, 1015, 1016, 1017, 1018, 1019, and 1021.
Related to images 1010, 1011, 1012, 1013, 1015, 1016, 1017, 1018, 1019, and 1021.
Andrew S. Bajer, University of Oregon, Eugene
View Media
6547: Cell Nucleus and Lipid Droplets
6547: Cell Nucleus and Lipid Droplets
A cell nucleus (blue) surrounded by lipid droplets (yellow). Exogenously expressed, S-tagged UBXD8 (green) recruits endogenous p97/VCP (red) to the surface of lipid droplets in oleate-treated HeLa cells. Nucleus stained with DAPI.
James Olzmann, University of California, Berkeley
View Media
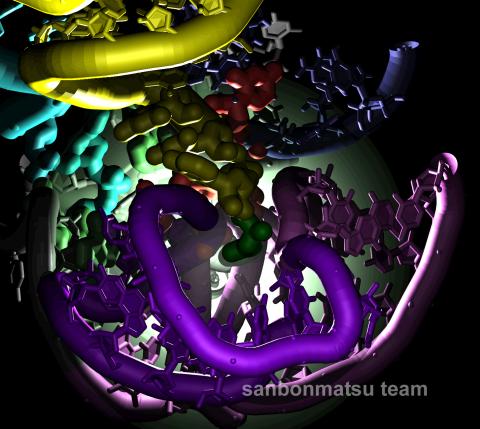
2336: Natural nanomachine in action
2336: Natural nanomachine in action
Using a supercomputer to simulate the movement of atoms in a ribosome, researchers looked into the core of this protein-making nanomachine and took snapshots. The picture shows an amino acid (green) being delivered by transfer RNA (yellow) into a corridor (purple) in the ribosome. In the corridor, a series of chemical reactions will string together amino acids to make a protein. The research project, which tracked the movement of more than 2.6 million atoms, was the largest computer simulation of a biological structure to date. The results shed light on the manufacturing of proteins and could aid the search for new antibiotics, which typically work by disabling the ribosomes of bacteria.
Kevin Sanbonmatsu, Los Alamos National Laboratory
View Media
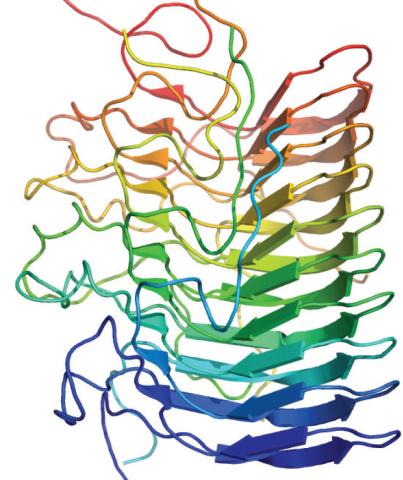
3542: Structure of amyloid-forming prion protein
3542: Structure of amyloid-forming prion protein
This structure from an amyloid-forming prion protein shows one way beta sheets can stack. Image originally appeared in a December 2012 PLOS Biology paper.
Douglas Fowler, University of Washington
View Media
3329: Spreading Cells- 02
3329: Spreading Cells- 02
Cells move forward with lamellipodia and filopodia supported by networks and bundles of actin filaments. Proper, controlled cell movement is a complex process. Recent research has shown that an actin-polymerizing factor called the Arp2/3 complex is the key component of the actin polymerization engine that drives amoeboid cell motility. ARPC3, a component of the Arp2/3 complex, plays a critical role in actin nucleation. In this photo, the ARPC3-/- fibroblast cells were fixed and stained with Alexa 546 phalloidin for F-actin (red), Arp2 (green), and DAPI to visualize the nucleus (blue). Arp2, a subunit of the Arp2/3 complex, is absent in the filopodi-like structures based leading edge of ARPC3-/- fibroblasts cells. Related to images 3328, 3330, 3331, 3332, and 3333.
Rong Li and Praveen Suraneni, Stowers Institute for Medical Research
View Media

2423: Protein map
2423: Protein map
Network diagram showing a map of protein-protein interactions in a yeast (Saccharomyces cerevisiae) cell. This cluster includes 78 percent of the proteins in the yeast proteome. The color of a node represents the phenotypic effect of removing the corresponding protein (red, lethal; green, nonlethal; orange, slow growth; yellow, unknown).
Hawoong Jeong, KAIST, Korea
View Media
5852: Optic nerve astrocytes
5852: Optic nerve astrocytes
Astrocytes in the cross section of a human optic nerve head
Tom Deerinck and Keunyoung (“Christine”) Kim, NCMIR
View Media
5767: Multivesicular bodies containing intralumenal vesicles assemble at the vacuole 3
5767: Multivesicular bodies containing intralumenal vesicles assemble at the vacuole 3
Collecting and transporting cellular waste and sorting it into recylable and nonrecylable pieces is a complex business in the cell. One key player in that process is the endosome, which helps collect, sort and transport worn-out or leftover proteins with the help of a protein assembly called the endosomal sorting complexes for transport (or ESCRT for short). These complexes help package proteins marked for breakdown into intralumenal vesicles, which, in turn, are enclosed in multivesicular bodies for transport to the places where the proteins are recycled or dumped. In this image, two multivesicular bodies (with yellow membranes) contain tiny intralumenal vesicles (with a diameter of only 25 nanometers; shown in red) adjacent to the cell's vacuole (in orange).
Scientists working with baker's yeast (Saccharomyces cerevisiae) study the budding inward of the limiting membrane (green lines on top of the yellow lines) into the intralumenal vesicles. This tomogram was shot with a Tecnai F-20 high-energy electron microscope, at 29,000x magnification, with a 0.7-nm pixel, ~4-nm resolution.
To learn more about endosomes, see the Biomedical Beat blog post The Cell’s Mailroom. Related to a microscopy photograph 5768 that was used to generate this illustration and a zoomed-out version 5769 of this illustration.
Scientists working with baker's yeast (Saccharomyces cerevisiae) study the budding inward of the limiting membrane (green lines on top of the yellow lines) into the intralumenal vesicles. This tomogram was shot with a Tecnai F-20 high-energy electron microscope, at 29,000x magnification, with a 0.7-nm pixel, ~4-nm resolution.
To learn more about endosomes, see the Biomedical Beat blog post The Cell’s Mailroom. Related to a microscopy photograph 5768 that was used to generate this illustration and a zoomed-out version 5769 of this illustration.
Matthew West and Greg Odorizzi, University of Colorado
View Media
2795: Anti-tumor drug ecteinascidin 743 (ET-743), structure without hydrogens 02
2795: Anti-tumor drug ecteinascidin 743 (ET-743), structure without hydrogens 02
Ecteinascidin 743 (ET-743, brand name Yondelis), was discovered and isolated from a sea squirt, Ecteinascidia turbinata, by NIGMS grantee Kenneth Rinehart at the University of Illinois. It was synthesized by NIGMS grantees E.J. Corey and later by Samuel Danishefsky. Multiple versions of this structure are available as entries 2790-2797.
Timothy Jamison, Massachusetts Institute of Technology
View Media
3788: Yeast cells pack a punch
3788: Yeast cells pack a punch
Although they are tiny, microbes that are growing in confined spaces can generate a lot of pressure. In this video, yeast cells grow in a small chamber called a microfluidic bioreactor. As the cells multiply, they begin to bump into and squeeze each other, resulting in periodic bursts of cells moving into different parts of the chamber. The continually growing cells also generate a lot of pressure--the researchers conducting these experiments found that the pressure generated by the cells can be almost five times higher than that in a car tire--about 150 psi, or 10 times the atmospheric pressure. Occasionally, this pressure even caused the small reactor to burst. By tracking the growth of the yeast or other cells and measuring the mechanical forces generated, scientists can simulate microbial growth in various places such as water pumps, sewage lines or catheters to learn how damage to these devices can be prevented. To learn more how researchers used small bioreactors to gauge the pressure generated by growing microbes, see this press release from UC Berkeley.
Oskar Hallatschek, UC Berkeley
View Media
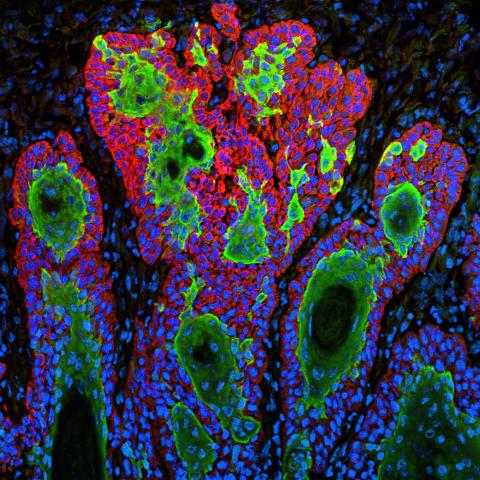
3628: Skin cancer cells (squamous cell carcinoma)
3628: Skin cancer cells (squamous cell carcinoma)
This image shows the uncontrolled growth of cells in squamous cell carcinoma, the second most common form of skin cancer. If caught early, squamous cell carcinoma is usually not life-threatening.
This image was part of the Life: Magnified exhibit that ran from June 3, 2014, to January 21, 2015, at Dulles International Airport.
This image was part of the Life: Magnified exhibit that ran from June 3, 2014, to January 21, 2015, at Dulles International Airport.
Markus Schober and Elaine Fuchs, The Rockefeller University
View Media
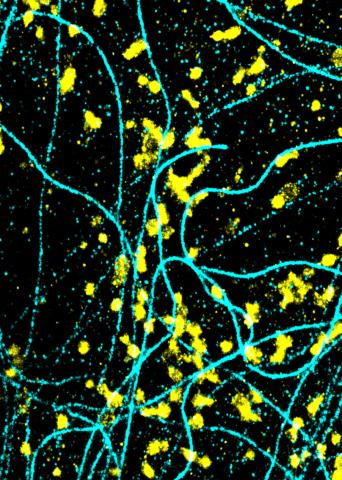
6889: Lysosomes and microtubules
6889: Lysosomes and microtubules
Lysosomes (yellow) and detyrosinated microtubules (light blue). Lysosomes are bubblelike organelles that take in molecules and use enzymes to break them down. Microtubules are strong, hollow fibers that provide structural support to cells. The researchers who took this image found that in epithelial cells, detyrosinated microtubules are a small subset of fibers, and they concentrate lysosomes around themselves. This image was captured using Stochastic Optical Reconstruction Microscopy (STORM).
Related to images 6890, 6891, and 6892.
Related to images 6890, 6891, and 6892.
Melike Lakadamyali, Perelman School of Medicine at the University of Pennsylvania.
View Media
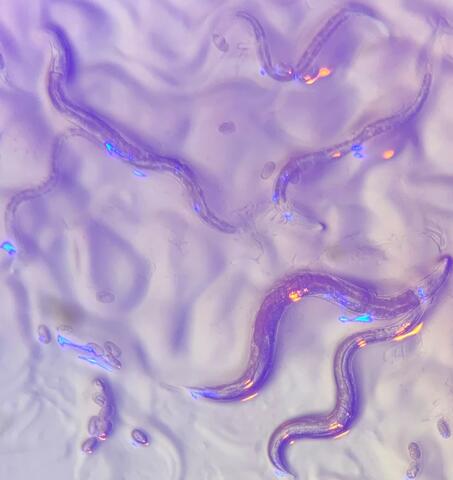
6750: C. elegans with blue and yellow lights in the background
6750: C. elegans with blue and yellow lights in the background
These microscopic roundworms, called Caenorhabditis elegans, lack eyes and the opsin proteins used by visual systems to detect colors. However, researchers found that the worms can still sense the color of light in a way that enables them to avoid pigmented toxins made by bacteria. This image was captured using a stereo microscope.
H. Robert Horvitz and Dipon Ghosh, Massachusetts Institute of Technology.
View Media
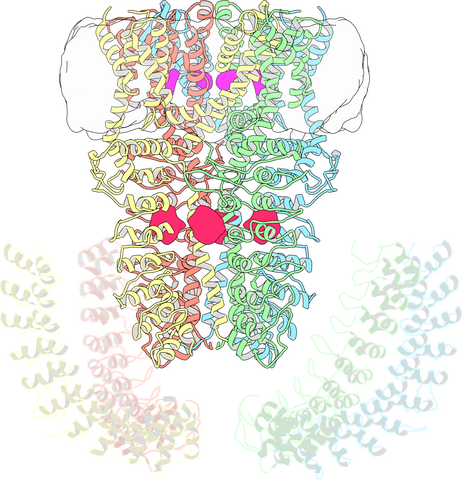
3747: Cryo-electron microscopy revealing the "wasabi receptor"
3747: Cryo-electron microscopy revealing the "wasabi receptor"
The TRPA1 protein is responsible for the burn you feel when you taste a bite of sushi topped with wasabi. Known therefore informally as the "wasabi receptor," this protein forms pores in the membranes of nerve cells that sense tastes or odors. Pungent chemicals like wasabi or mustard oil cause the pores to open, which then triggers a tingling or burn on our tongue. This receptor also produces feelings of pain in response to chemicals produced within our own bodies when our tissues are damaged or inflamed. Researchers used cryo-EM to reveal the structure of the wasabi receptor at a resolution of about 4 angstroms (a credit card is about 8 million angstroms thick). This detailed structure can help scientists understand both how we feel pain and how we can limit it by developing therapies to block the receptor. For more on cryo-EM see the blog post Cryo-Electron Microscopy Reveals Molecules in Ever Greater Detail.
Jean-Paul Armache, UCSF
View Media
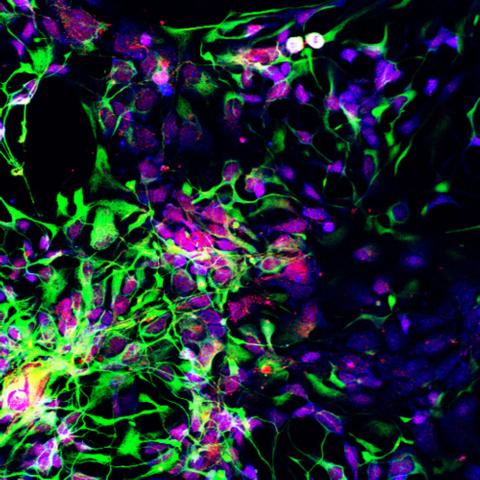
3280: Motor neuron progenitors derived from human ES cells
3280: Motor neuron progenitors derived from human ES cells
Motor neuron progenitors (green) were derived from human embryonic stem cells. Image and caption information courtesy of the California Institute for Regenerative Medicine.
Hans Keirstead lab, University of California, Irvine, via CIRM
View Media
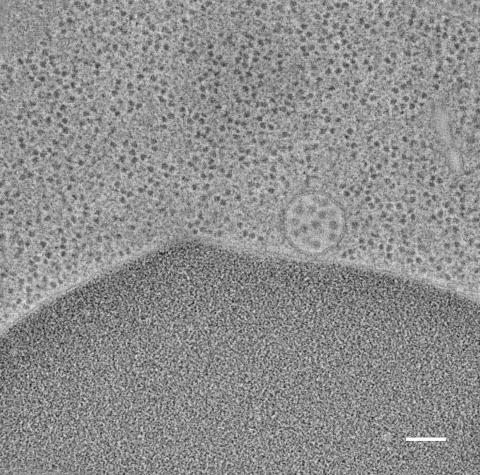
5768: Multivesicular bodies containing intralumenal vesicles assemble at the vacuole 2
5768: Multivesicular bodies containing intralumenal vesicles assemble at the vacuole 2
Collecting and transporting cellular waste and sorting it into recylable and nonrecylable pieces is a complex business in the cell. One key player in that process is the endosome, which helps collect, sort and transport worn-out or leftover proteins with the help of a protein assembly called the endosomal sorting complexes for transport (or ESCRT for short). These complexes help package proteins marked for breakdown into intralumenal vesicles, which, in turn, are enclosed in multivesicular bodies for transport to the places where the proteins are recycled or dumped. In this image, a multivesicular body (the round structure slightly to the right of center) contain tiny intralumenal vesicles (with a diameter of only 25 nanometers; the round specks inside the larger round structure) adjacent to the cell's vacuole (below the multivesicular body, shown in darker and more uniform gray).
Scientists working with baker's yeast (Saccharomyces cerevisiae) study the budding inward of the limiting membrane (green lines on top of the yellow lines) into the intralumenal vesicles. This tomogram was shot with a Tecnai F-20 high-energy electron microscope, at 29,000x magnification, with a 0.7-nm pixel, ~4-nm resolution.
To learn more about endosomes, see the Biomedical Beat blog post The Cell’s Mailroom. Related to a color-enhanced version 5767 and image 5769.
Scientists working with baker's yeast (Saccharomyces cerevisiae) study the budding inward of the limiting membrane (green lines on top of the yellow lines) into the intralumenal vesicles. This tomogram was shot with a Tecnai F-20 high-energy electron microscope, at 29,000x magnification, with a 0.7-nm pixel, ~4-nm resolution.
To learn more about endosomes, see the Biomedical Beat blog post The Cell’s Mailroom. Related to a color-enhanced version 5767 and image 5769.
Matthew West and Greg Odorizzi, University of Colorado
View Media
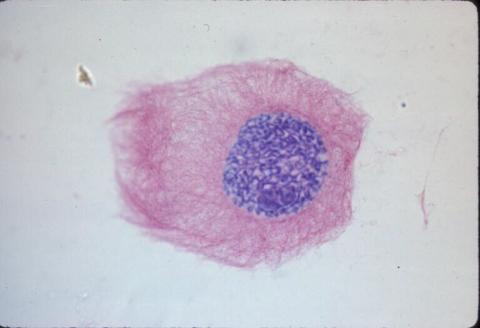
1013: Lily mitosis 03
1013: Lily mitosis 03
A light microscope image of a cell from the endosperm of an African globe lily (Scadoxus katherinae). This is one frame of a time-lapse sequence that shows cell division in action. The lily is considered a good organism for studying cell division because its chromosomes are much thicker and easier to see than human ones. Staining shows microtubules in red and chromosomes in blue.
Related to images 1010, 1011, 1012, 1014, 1015, 1016, 1017, 1018, 1019, and 1021.
Related to images 1010, 1011, 1012, 1014, 1015, 1016, 1017, 1018, 1019, and 1021.
Andrew S. Bajer, University of Oregon, Eugene
View Media

7013: An adult Hawaiian bobtail squid
7013: An adult Hawaiian bobtail squid
An adult female Hawaiian bobtail squid, Euprymna scolopes, with its mantle cavity exposed from the underside. Some internal organs are visible, including the two lobes of the light organ that contains bioluminescent bacteria, Vibrio fischeri. The light organ includes accessory tissues like an ink sac (black) that serves as a shutter, and a silvery reflector that directs the light out of the underside of the animal.
Margaret J. McFall-Ngai, Carnegie Institution for Science/California Institute of Technology, and Edward G. Ruby, California Institute of Technology.
View Media
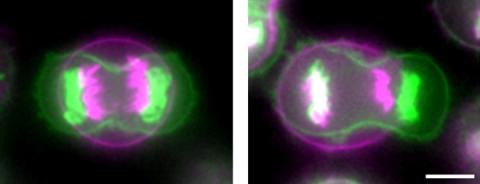
3648: Symmetrically and asymmetrically elongating cells
3648: Symmetrically and asymmetrically elongating cells
Merged fluorescent images of symmetrically (left) or asymmetrically (right) elongating HeLa cells at the end of early anaphase (magenta) and late anaphase (green). Chromosomes and cortical actin are visualized by expressing mCherry-histone H2B and Lifeact-mCherry. Scale bar, 10µm. See the PubMed abstract of this research.
Tomomi Kiyomitsu and Iain M. Cheeseman, Whitehead Institute for Biomedical Research
View Media
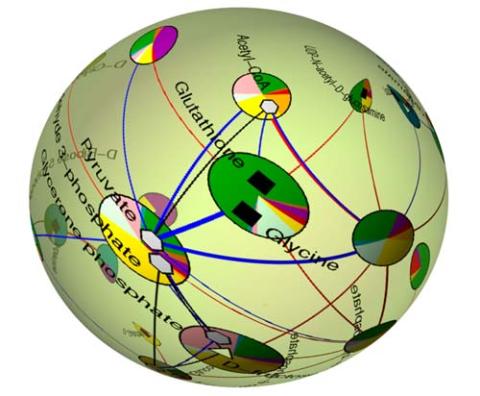
2331: Statistical cartography
2331: Statistical cartography
Like a world of its own, this sphere represents all the known chemical reactions in the E. coli bacterium. The colorful circles on the surface symbolize sets of densely interconnected reactions. The lines between the circles show additional connecting reactions. The shapes inside the circles are landmark molecules, like capital cities on a map, that either act as hubs for many groups of reactions, are highly conserved among species, or both. Molecules that connect far-flung reactions on the sphere are much more conserved during evolution than molecules that connect reactions within a single circle. This statistical cartography could reveal insights about other complex systems, such as protein interactions and gene regulation networks.
Luis A. Nunes Amaral, Northwestern University
View Media
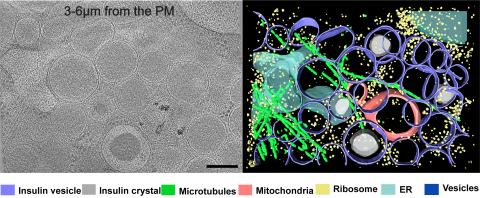
6607: Cryo-ET cell cross-section visualizing insulin vesicles
6607: Cryo-ET cell cross-section visualizing insulin vesicles
On the left, a cross-section slice of a rat pancreas cell captured using cryo-electron tomography (cryo-ET). On the right, a color-coded, 3D version of the image highlighting cell structures. Visible features include insulin vesicles (purple rings), insulin crystals (gray circles), microtubules (green rods), ribosomes (small yellow circles). The black line at the bottom right of the left image represents 200 nm. Related to image 6608.
Xianjun Zhang, University of Southern California.
View Media
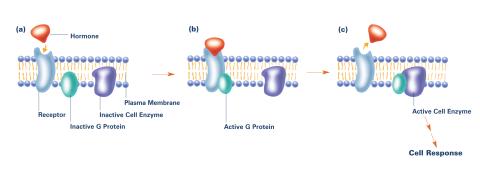
2538: G switch (with labels and stages)
2538: G switch (with labels and stages)
The G switch allows our bodies to respond rapidly to hormones. G proteins act like relay batons to pass messages from circulating hormones into cells. A hormone (red) encounters a receptor (blue) in the membrane of a cell. Next, a G protein (green) becomes activated and makes contact with the receptor to which the hormone is attached. Finally, the G protein passes the hormone's message to the cell by switching on a cell enzyme (purple) that triggers a response. See image 2536 and 2537 for other versions of this image. Featured in Medicines By Design.
Crabtree + Company
View Media
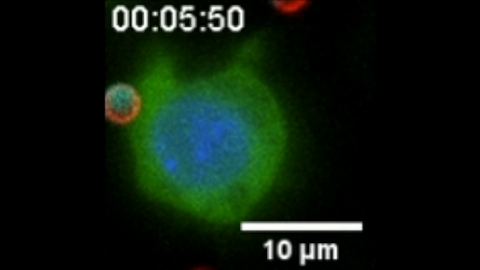
6801: “Two-faced” Janus particle activating a macrophage
6801: “Two-faced” Janus particle activating a macrophage
A macrophage—a type of immune cell that engulfs invaders—“eats” and is activated by a “two-faced” Janus particle. The particle is called “two-faced” because each of its two hemispheres is coated with a different type of molecule, shown here in red and cyan. During macrophage activation, a transcription factor tagged with a green fluorescence protein (NF-κB) gradually moves from the cell’s cytoplasm into its nucleus and causes DNA transcription. The distribution of molecules on “two-faced” Janus particles can be altered to control the activation of immune cells. Details on this “geometric manipulation” strategy can be found in the Proceedings of the National Academy of Sciences paper "Geometrical reorganization of Dectin-1 and TLR2 on single phagosomes alters their synergistic immune signaling" by Li et al. and the Scientific Reports paper "Spatial organization of FcγR and TLR2/1 on phagosome membranes differentially regulates their synergistic and inhibitory receptor crosstalk" by Li et al. This video was captured using epi-fluorescence microscopy.
Related to video 6800.
Related to video 6800.
Yan Yu, Indiana University, Bloomington.
View Media
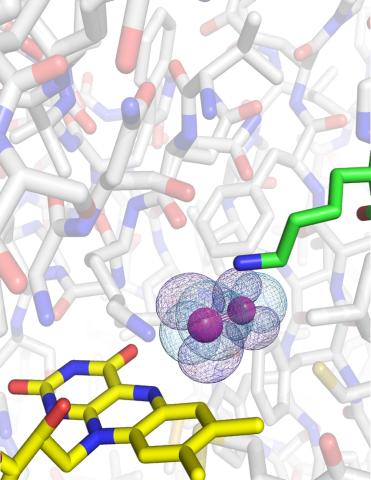
3411: O2 reacting with a flavin-dependent enzyme
3411: O2 reacting with a flavin-dependent enzyme
Department of Biological Chemistry, University of Michigan
View Media
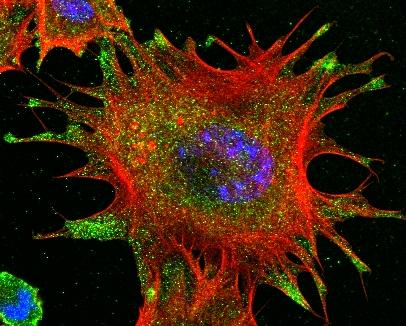
3331: mDia1 antibody staining- 02
3331: mDia1 antibody staining- 02
Cells move forward with lamellipodia and filopodia supported by networks and bundles of actin filaments. Proper, controlled cell movement is a complex process. Recent research has shown that an actin-polymerizing factor called the Arp2/3 complex is the key component of the actin polymerization engine that drives amoeboid cell motility. ARPC3, a component of the Arp2/3 complex, plays a critical role in actin nucleation. In this photo, the ARPC3-/- fibroblast cells were fixed and stained with Alexa 546 phalloidin for F-actin (red), mDia1 (green), and DAPI to visualize the nucleus (blue). In ARPC3-/- fibroblast cells, mDia1 is localized at the tips of the filopodia-like structures. Related to images 3328, 3329, 3330, 3332, and 3333.
Rong Li and Praveen Suraneni, Stowers Institute for Medical Research
View Media
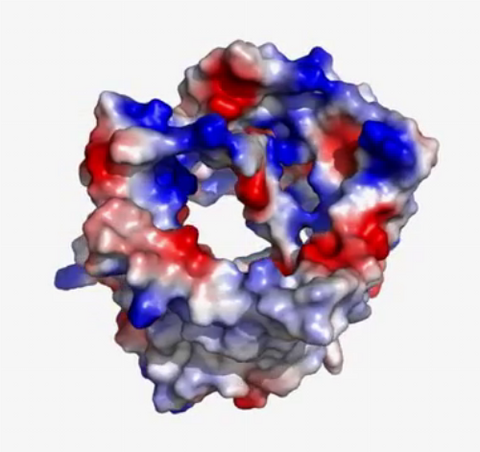
2571: VDAC video 02
2571: VDAC video 02
This video shows the structure of the pore-forming protein VDAC-1 from humans. This molecule mediates the flow of products needed for metabolism--in particular the export of ATP--across the outer membrane of mitochondria, the power plants for eukaryotic cells. VDAC-1 is involved in metabolism and the self-destruction of cells--two biological processes central to health.
Related to videos 2570 and 2572.
Related to videos 2570 and 2572.
Gerhard Wagner, Harvard Medical School
View Media
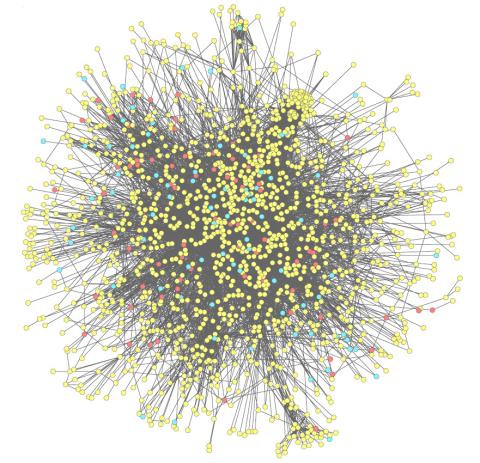
2743: Molecular interactions
2743: Molecular interactions
This network map shows molecular interactions (yellow) associated with a congenital condition that causes heart arrhythmias and the targets for drugs that alter these interactions (red and blue).
Ravi Iyengar, Mount Sinai School of Medicine
View Media

1287: Mitochondria
1287: Mitochondria
Bean-shaped mitochondria are cells' power plants. These organelles have their own DNA and replicate independently. The highly folded inner membranes are the site of energy generation.
Judith Stoffer
View Media
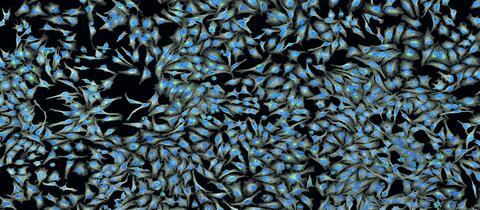
5761: A panorama view of cells
5761: A panorama view of cells
This photograph shows a panoramic view of HeLa cells, a cell line many researchers use to study a large variety of important research questions. The cells' nuclei containing the DNA are stained in blue and the cells' cytoskeletons in gray.
Tom Deerinck, National Center for Microscopy and Imaging Research
View Media
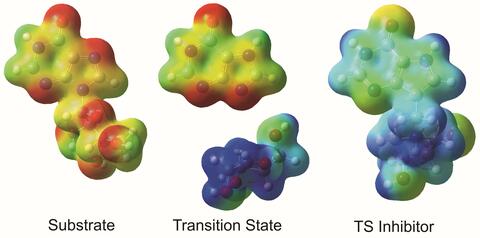
3429: Enzyme transition states
3429: Enzyme transition states
The molecule on the left is an electrostatic potential map of the van der Waals surface of the transition state for human purine nucleoside phosphorylase. The colors indicate the electron density at any position of the molecule. Red indicates electron-rich regions with negative charge and blue indicates electron-poor regions with positive charge. The molecule on the right is called DADMe-ImmH. It is a chemically stable analogue of the transition state on the left. It binds to the enzyme millions of times tighter than the substrate. This inhibitor is in human clinical trials for treating patients with gout. This image appears in Figure 4, Schramm, V.L. (2011) Annu. Rev. Biochem. 80:703-732.
Vern Schramm, Albert Einstein College of Medicine of Yeshiva University
View Media
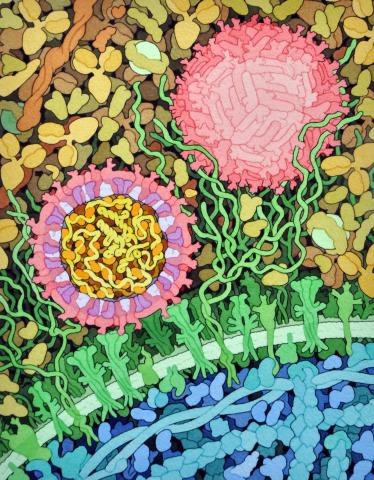
6998: Zika virus
6998: Zika virus
Zika virus is shown in cross section at center left. On the outside, it includes envelope protein (red) and membrane protein (magenta) embedded in a lipid membrane (light purple). Inside, the RNA genome (yellow) is associated with capsid proteins (orange). The viruses are shown interacting with receptors on the cell surface (green) and are surrounded by blood plasma molecules at the top.
Amy Wu and Christine Zardecki, RCSB Protein Data Bank.
View Media
6553: Floral pattern in a mixture of two bacterial species, Acinetobacter baylyi and Escherichia coli, grown on a semi-solid agar for 48 hours (photo 1)
6553: Floral pattern in a mixture of two bacterial species, Acinetobacter baylyi and Escherichia coli, grown on a semi-solid agar for 48 hours (photo 1)
Floral pattern emerging as two bacterial species, motile Acinetobacter baylyi (red) and non-motile Escherichia coli (green), are grown together for 48 hours on 1% agar surface from a small inoculum in the center of a Petri dish.
See 6557 for a photo of this process at 24 hours on 0.75% agar surface.
See 6555 for another photo of this process at 48 hours on 1% agar surface.
See 6556 for a photo of this process at 72 hours on 0.5% agar surface.
See 6550 for a video of this process.
See 6557 for a photo of this process at 24 hours on 0.75% agar surface.
See 6555 for another photo of this process at 48 hours on 1% agar surface.
See 6556 for a photo of this process at 72 hours on 0.5% agar surface.
See 6550 for a video of this process.
L. Xiong et al, eLife 2020;9: e48885
View Media
6389: Red and white blood cells in the lung
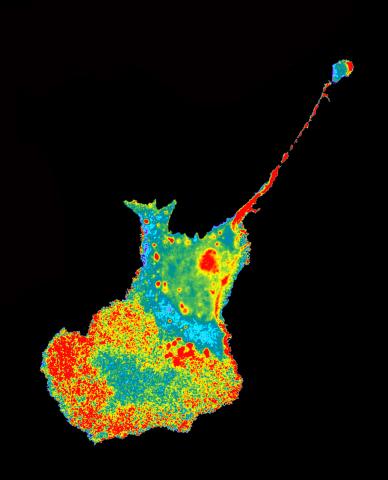
2454: Seeing signaling protein activation in cells 04
2454: Seeing signaling protein activation in cells 04
Cdc42, a member of the Rho family of small guanosine triphosphatase (GTPase) proteins, regulates multiple cell functions, including motility, proliferation, apoptosis, and cell morphology. In order to fulfill these diverse roles, the timing and location of Cdc42 activation must be tightly controlled. Klaus Hahn and his research group use special dyes designed to report protein conformational changes and interactions, here in living neutrophil cells. Warmer colors in this image indicate higher levels of activation. Cdc42 looks to be activated at cell protrusions.
Related to images 2451, 2452, and 2453.
Related to images 2451, 2452, and 2453.
Klaus Hahn, University of North Carolina, Chapel Hill Medical School
View Media
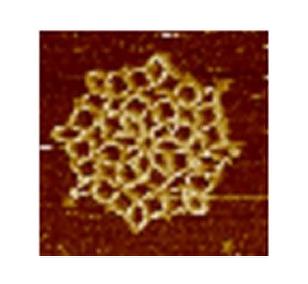
3724: Snowflake DNA origami
3724: Snowflake DNA origami
An atomic force microscopy image shows DNA folded into an intricate, computer-designed structure. The image is featured on Biomedical Beat blog post Cool Images: A Holiday-Themed Collection. For more background on DNA origami, see Cool Image: DNA Origami. See also related image 3690.
Hao Yan, Arizona State University
View Media
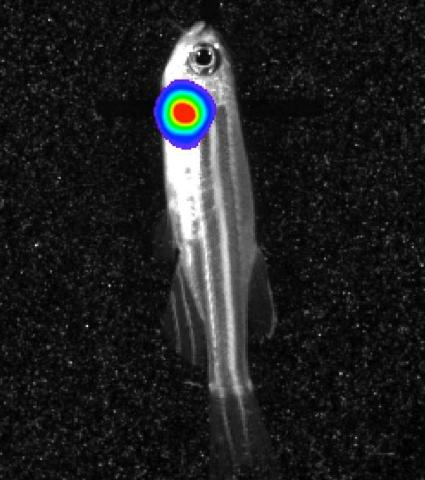
3558: Bioluminescent imaging in adult zebrafish - lateral view
3558: Bioluminescent imaging in adult zebrafish - lateral view
Luciferase-based imaging enables visualization and quantification of internal organs and transplanted cells in live adult zebrafish. In this image, a cardiac muscle-restricted promoter drives firefly luciferase expression (lateral view).
For imagery of both the lateral and overhead view go to 3556.
For imagery of the overhead view go to 3557.
For more information about the illumated area go to 3559.
For imagery of both the lateral and overhead view go to 3556.
For imagery of the overhead view go to 3557.
For more information about the illumated area go to 3559.
Kenneth Poss, Duke University
View Media
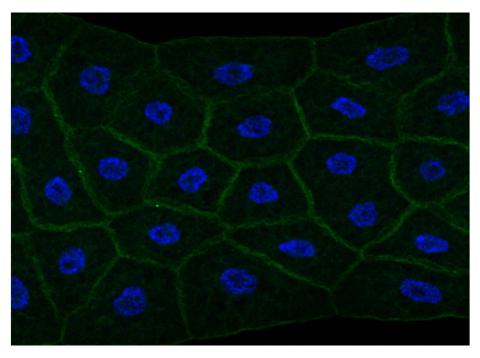
2757: Draper, shown in the fatbody of a Drosophila melanogaster larva
2757: Draper, shown in the fatbody of a Drosophila melanogaster larva
The fly fatbody is a nutrient storage and mobilization organ akin to the mammalian liver. The engulfment receptor Draper (green) is located at the cell surface of fatbody cells. The cell nuclei are shown in blue.
Christina McPhee and Eric Baehrecke, University of Massachusetts Medical School
View Media

6769: Culex quinquefasciatus mosquito larva
6769: Culex quinquefasciatus mosquito larva
A mosquito larva with genes edited by CRISPR. The red-orange glow is a fluorescent protein used to track the edits. This species of mosquito, Culex quinquefasciatus, can transmit West Nile virus, Japanese encephalitis virus, and avian malaria, among other diseases. The researchers who took this image developed a gene-editing toolkit for Culex quinquefasciatus that could ultimately help stop the mosquitoes from spreading pathogens. The work is described in the Nature Communications paper "Optimized CRISPR tools and site-directed transgenesis towards gene drive development in Culex quinquefasciatus mosquitoes" by Feng et al. Related to image 6770 and video 6771.
Valentino Gantz, University of California, San Diego.
View Media
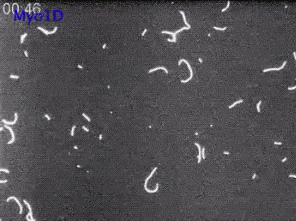
6562: Drosophila (fruit fly) myosin 1D motility assay
6562: Drosophila (fruit fly) myosin 1D motility assay
Actin gliding powered by myosin 1D. Note the counterclockwise motion of the gliding actin filaments.
Serapion Pyrpassopoulos and E. Michael Ostap, University of Pennsylvania
View Media