Switch to List View
Image and Video Gallery
This is a searchable collection of scientific photos, illustrations, and videos. The images and videos in this gallery are licensed under Creative Commons Attribution Non-Commercial ShareAlike 3.0. This license lets you remix, tweak, and build upon this work non-commercially, as long as you credit and license your new creations under identical terms.
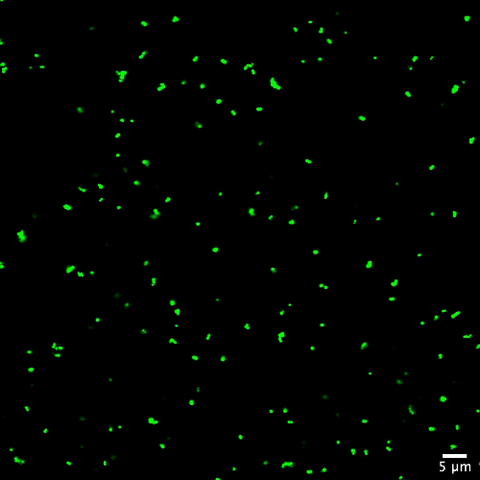
6805: Staphylococcus aureus aggregating upon contact with synovial fluid
6805: Staphylococcus aureus aggregating upon contact with synovial fluid
Staphylococcus aureus bacteria (green) grouping together upon contact with synovial fluid—a viscous substance found in joints. The formation of groups can help protect the bacteria from immune system defenses and from antibiotics, increasing the likelihood of an infection. This video is a 1-hour time lapse and was captured using a confocal laser scanning microscope.
More information about the research that produced this video can be found in the Journal of Bacteriology paper "In Vitro Staphylococcal Aggregate Morphology and Protection from Antibiotics Are Dependent on Distinct Mechanisms Arising from Postsurgical Joint Components and Fluid Motion" by Staats et al.
Related to images 6803 and 6804.
More information about the research that produced this video can be found in the Journal of Bacteriology paper "In Vitro Staphylococcal Aggregate Morphology and Protection from Antibiotics Are Dependent on Distinct Mechanisms Arising from Postsurgical Joint Components and Fluid Motion" by Staats et al.
Related to images 6803 and 6804.
Paul Stoodley, The Ohio State University.
View Media

2411: Fungal lipase (2)
2411: Fungal lipase (2)
Crystals of fungal lipase protein created for X-ray crystallography, which can reveal detailed, three-dimensional protein structures.
Alex McPherson, University of California, Irvine
View Media
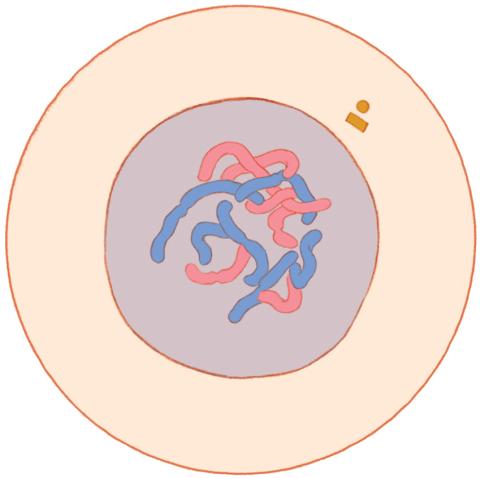
1316: Mitosis - interphase
1316: Mitosis - interphase
A cell in interphase, at the start of mitosis: Chromosomes duplicate, and the copies remain attached to each other. Mitosis is responsible for growth and development, as well as for replacing injured or worn out cells throughout the body. For simplicity, mitosis is illustrated here with only six chromosomes.
Judith Stoffer
View Media
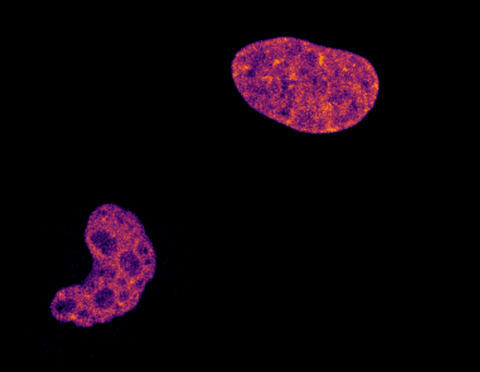
6790: Cell division and cell death
6790: Cell division and cell death
Two cells over a 2-hour period. The one on the bottom left goes through programmed cell death, also known as apoptosis. The one on the top right goes through cell division, also called mitosis. This video was captured using a confocal microscope.
Dylan T. Burnette, Vanderbilt University School of Medicine.
View Media
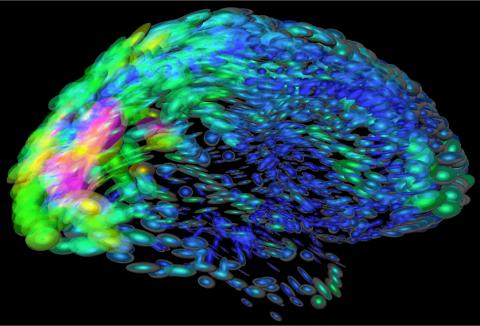
2419: Mapping brain differences
2419: Mapping brain differences
This image of the human brain uses colors and shapes to show neurological differences between two people. The blurred front portion of the brain, associated with complex thought, varies most between the individuals. The blue ovals mark areas of basic function that vary relatively little. Visualizations like this one are part of a project to map complex and dynamic information about the human brain, including genes, enzymes, disease states, and anatomy. The brain maps represent collaborations between neuroscientists and experts in math, statistics, computer science, bioinformatics, imaging, and nanotechnology.
Arthur Toga, University of California, Los Angeles
View Media
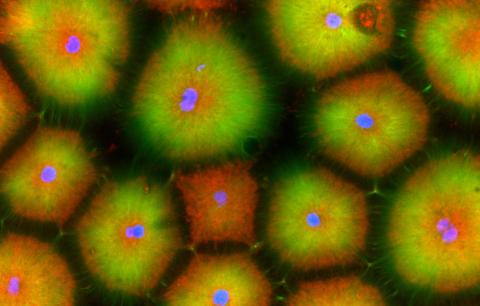
6592: Cell-like compartments from frog eggs 5
6592: Cell-like compartments from frog eggs 5
Cell-like compartments that spontaneously emerged from scrambled frog eggs, with nuclei (blue) from frog sperm. Endoplasmic reticulum (red) and microtubules (green) are also visible. Image created using confocal microscopy.
For more photos of cell-like compartments from frog eggs view: 6584, 6585, 6586, 6591, and 6593.
For videos of cell-like compartments from frog eggs view: 6587, 6588, 6589, and 6590.
Xianrui Cheng, Stanford University School of Medicine.
View Media
6389: Red and white blood cells in the lung
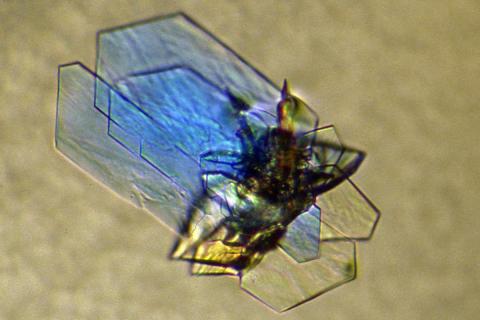
2410: DNase
2410: DNase
Crystals of DNase protein created for X-ray crystallography, which can reveal detailed, three-dimensional protein structures.
Alex McPherson, University of California, Irvine
View Media
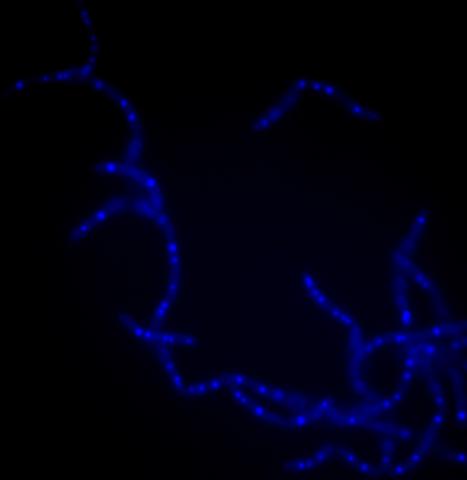
3525: Bacillus anthracis being killed
3525: Bacillus anthracis being killed
Bacillus anthracis (anthrax) cells being killed by a fluorescent trans-translation inhibitor, which disrupts bacterial protein synthesis. The inhibitor is naturally fluorescent and looks blue when it is excited by ultraviolet light in the microscope. This is a color version of Image 3481.
Kenneth Keiler, Penn State University
View Media
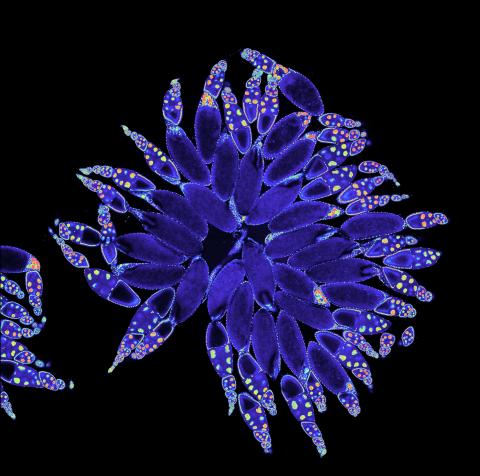
3607: Fruit fly ovary
3607: Fruit fly ovary
A fruit fly ovary, shown here, contains as many as 20 eggs. Fruit flies are not merely tiny insects that buzz around overripe fruit—they are a venerable scientific tool. Research on the flies has shed light on many aspects of human biology, including biological rhythms, learning, memory, and neurodegenerative diseases. Another reason fruit flies are so useful in a lab (and so successful in fruit bowls) is that they reproduce rapidly. About three generations can be studied in a single month.
Related to image 3656. This image was part of the Life: Magnified exhibit that ran from June 3, 2014, to January 21, 2015, at Dulles International Airport.
Related to image 3656. This image was part of the Life: Magnified exhibit that ran from June 3, 2014, to January 21, 2015, at Dulles International Airport.
Denise Montell, Johns Hopkins University and University of California, Santa Barbara
View Media
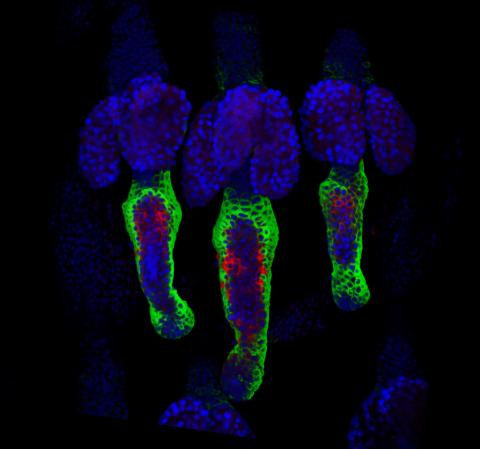
3497: Wound healing in process
3497: Wound healing in process
Wound healing requires the action of stem cells. In mice that lack the Sept2/ARTS gene, stem cells involved in wound healing live longer and wounds heal faster and more thoroughly than in normal mice. This confocal microscopy image from a mouse lacking the Sept2/ARTS gene shows a tail wound in the process of healing. See more information in the article in Science.
Related to images 3498 and 3500.
Related to images 3498 and 3500.
Hermann Steller, Rockefeller University
View Media

2345: Magnesium transporter protein from E. faecalis
2345: Magnesium transporter protein from E. faecalis
Structure of a magnesium transporter protein from an antibiotic-resistant bacterium (Enterococcus faecalis) found in the human gut. Featured as one of the June 2007 Protein Sructure Initiative Structures of the Month.
New York Structural GenomiX Consortium
View Media
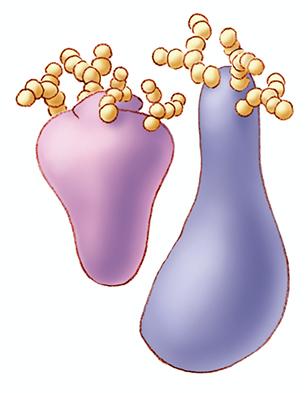
1270: Glycoproteins
1270: Glycoproteins
About half of all human proteins include chains of sugar molecules that are critical for the proteins to function properly. Appears in the NIGMS booklet Inside the Cell.
Judith Stoffer
View Media
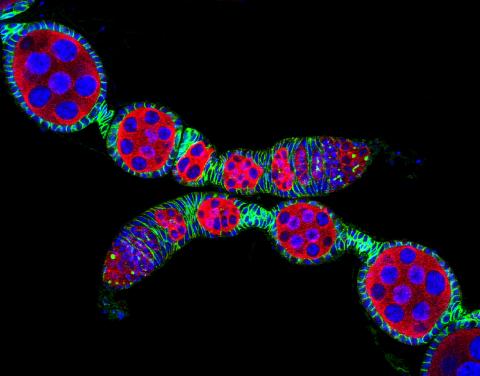
5772: Confocal microscopy image of two Drosophila ovarioles
5772: Confocal microscopy image of two Drosophila ovarioles
Ovarioles in female insects are tubes in which egg cells (called oocytes) form at one end and complete their development as they reach the other end of the tube. This image, taken with a confocal microscope, shows ovarioles in a very popular lab animal, the fruit fly Drosophila. The basic structure of ovarioles supports very rapid egg production, with some insects (like termites) producing several thousand eggs per day. Each insect ovary typically contains four to eight ovarioles, but this number varies widely depending on the insect species.
Scientists use insect ovarioles, for example, to study the basic processes that help various insects, including those that cause disease (like some mosquitos and biting flies), reproduce very quickly.
Scientists use insect ovarioles, for example, to study the basic processes that help various insects, including those that cause disease (like some mosquitos and biting flies), reproduce very quickly.
2004 Olympus BioScapes Competition
View Media

6356: H1N1 Influenza Virus
6356: H1N1 Influenza Virus
Related to image 6355.
Dr. Rommie Amaro, University of California, San Diego
View Media
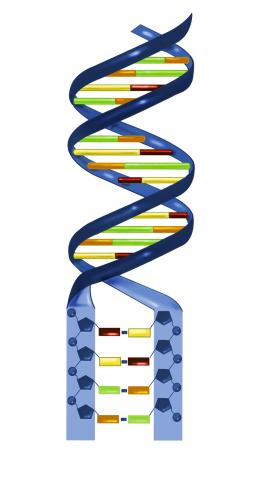
2541: Nucleotides make up DNA
2541: Nucleotides make up DNA
DNA consists of two long, twisted chains made up of nucleotides. Each nucleotide contains one base, one phosphate molecule, and the sugar molecule deoxyribose. The bases in DNA nucleotides are adenine, thymine, cytosine, and guanine. See image 2542 for a labeled version of this illustration. Featured in The New Genetics.
Crabtree + Company
View Media
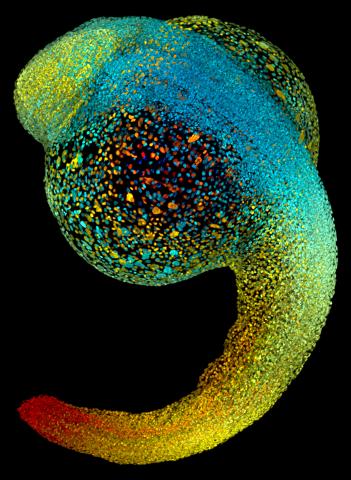
3644: Zebrafish embryo
3644: Zebrafish embryo
Just 22 hours after fertilization, this zebrafish embryo is already taking shape. By 36 hours, all of the major organs will have started to form. The zebrafish's rapid growth and see-through embryo make it ideal for scientists studying how organs develop.
This image was part of the Life: Magnified exhibit that ran from June 3, 2014, to January 21, 2015, at Dulles International Airport.
This image was part of the Life: Magnified exhibit that ran from June 3, 2014, to January 21, 2015, at Dulles International Airport.
Philipp Keller, Bill Lemon, Yinan Wan, and Kristin Branson, Janelia Farm Research Campus, Howard Hughes Medical Institute, Ashburn, Va.
View Media
6776: Tracking cells in a gastrulating zebrafish embryo
6776: Tracking cells in a gastrulating zebrafish embryo
During development, a zebrafish embryo is transformed from a ball of cells into a recognizable body plan by sweeping convergence and extension cell movements. This process is called gastrulation. Each line in this video represents the movement of a single zebrafish embryo cell over the course of 3 hours. The video was created using time-lapse confocal microscopy. Related to image 6775.
Liliana Solnica-Krezel, Washington University School of Medicine in St. Louis.
View Media
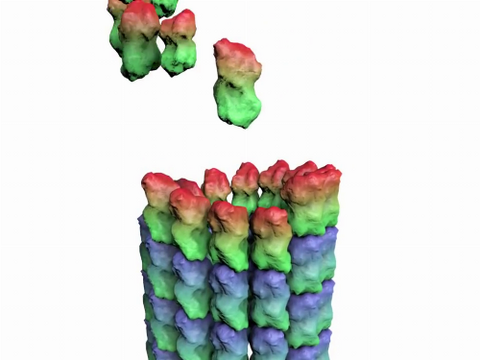
3650: How a microtubule builds and deconstructs
3650: How a microtubule builds and deconstructs
A microtubule, part of the cell's skeleton, builds and deconstructs.
View Media
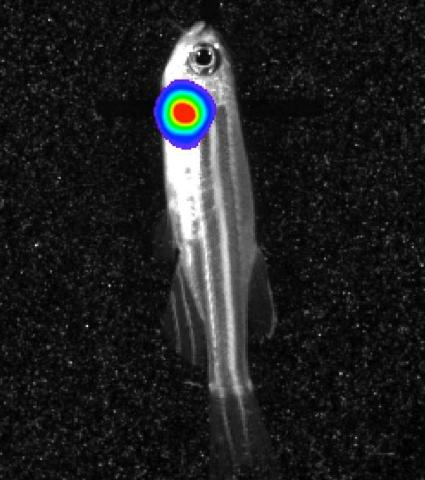
3558: Bioluminescent imaging in adult zebrafish - lateral view
3558: Bioluminescent imaging in adult zebrafish - lateral view
Luciferase-based imaging enables visualization and quantification of internal organs and transplanted cells in live adult zebrafish. In this image, a cardiac muscle-restricted promoter drives firefly luciferase expression (lateral view).
For imagery of both the lateral and overhead view go to 3556.
For imagery of the overhead view go to 3557.
For more information about the illumated area go to 3559.
For imagery of both the lateral and overhead view go to 3556.
For imagery of the overhead view go to 3557.
For more information about the illumated area go to 3559.
Kenneth Poss, Duke University
View Media
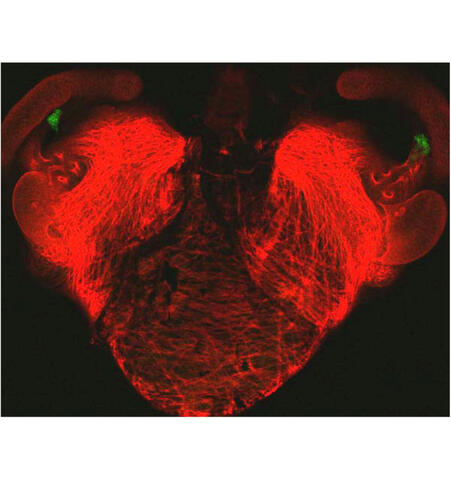
7018: Bacterial cells aggregating above the light organ of the Hawaiian bobtail squid
7018: Bacterial cells aggregating above the light organ of the Hawaiian bobtail squid
A light organ (~0.5 mm across) of a juvenile Hawaiian bobtail squid, Euprymna scolopes. Movement of cilia on the surface of the organ aggregates bacterial symbionts (green) into two areas above sets of pores that lead to interior crypts. This image was taken using a confocal fluorescence microscope.
Related to images 7016, 7017, 7019, and 7020.
Related to images 7016, 7017, 7019, and 7020.
Margaret J. McFall-Ngai, Carnegie Institution for Science/California Institute of Technology, and Edward G. Ruby, California Institute of Technology.
View Media
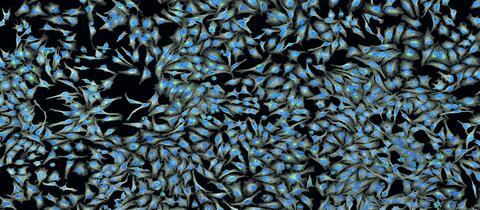
5761: A panorama view of cells
5761: A panorama view of cells
This photograph shows a panoramic view of HeLa cells, a cell line many researchers use to study a large variety of important research questions. The cells' nuclei containing the DNA are stained in blue and the cells' cytoskeletons in gray.
Tom Deerinck, National Center for Microscopy and Imaging Research
View Media

7012: Adult Hawaiian bobtail squid burying in the sand
7012: Adult Hawaiian bobtail squid burying in the sand
Each morning, the nocturnal Hawaiian bobtail squid, Euprymna scolopes, hides from predators by digging into the sand. At dusk, it leaves the sand again to hunt.
Related to image 7010 and 7011.
Related to image 7010 and 7011.
Margaret J. McFall-Ngai, Carnegie Institution for Science/California Institute of Technology, and Edward G. Ruby, California Institute of Technology.
View Media
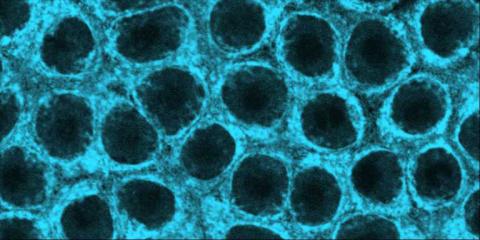
2324: Movements of myosin
2324: Movements of myosin
Inside the fertilized egg cell of a fruit fly, we see a type of myosin (related to the protein that helps muscles contract) made to glow by attaching a fluorescent protein. After fertilization, the myosin proteins are distributed relatively evenly near the surface of the embryo. The proteins temporarily vanish each time the cells' nuclei--initially buried deep in the cytoplasm--divide. When the multiplying nuclei move to the surface, they shift the myosin, producing darkened holes. The glowing myosin proteins then gather, contract, and start separating the nuclei into their own compartments.
Victoria Foe, University of Washington
View Media
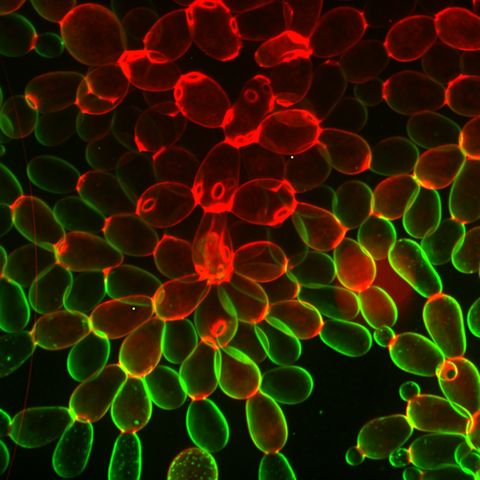
6969: Snowflake yeast 1
6969: Snowflake yeast 1
Multicellular yeast called snowflake yeast that researchers created through many generations of directed evolution from unicellular yeast. Stained cell membranes (green) and cell walls (red) reveal the connections between cells. Younger cells take up more cell membrane stain, while older cells take up more cell wall stain, leading to the color differences seen here. This image was captured using spinning disk confocal microscopy.
Related to images 6970 and 6971.
Related to images 6970 and 6971.
William Ratcliff, Georgia Institute of Technology.
View Media
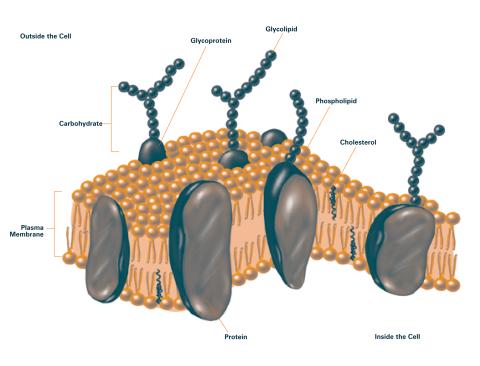
2524: Plasma membrane (with labels)
2524: Plasma membrane (with labels)
The plasma membrane is a cell's protective barrier. See image 2523 for an unlabeled version of this illustration. Featured in The Chemistry of Health.
Crabtree + Company
View Media
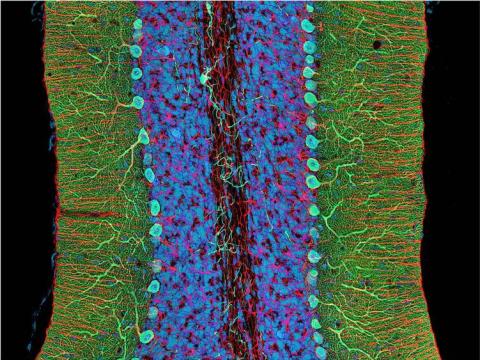
3371: Mouse cerebellum close-up
3371: Mouse cerebellum close-up
The cerebellum is the brain's locomotion control center. Every time you shoot a basketball, tie your shoe or chop an onion, your cerebellum fires into action. Found at the base of your brain, the cerebellum is a single layer of tissue with deep folds like an accordion. People with damage to this region of the brain often have difficulty with balance, coordination and fine motor skills. For a lower magnification, see image 3639.
This image was part of the Life: Magnified exhibit that ran from June 3, 2014, to January 21, 2015, at Dulles International Airport.
This image was part of the Life: Magnified exhibit that ran from June 3, 2014, to January 21, 2015, at Dulles International Airport.
National Center for Microscopy and Imaging Research (NCMIR)
View Media
2438: Hydra 02
2438: Hydra 02
Hydra magnipapillata is an invertebrate animal used as a model organism to study developmental questions, for example the formation of the body axis.
Hiroshi Shimizu, National Institute of Genetics in Mishima, Japan
View Media
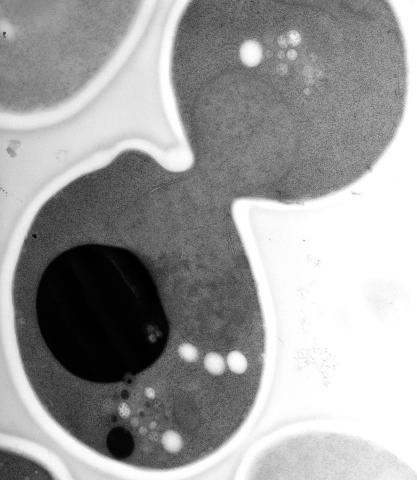
5770: EM of yeast cell division
5770: EM of yeast cell division
Cell division is an incredibly coordinated process. It not only ensures that the new cells formed during this event have a full set of chromosomes, but also that they are endowed with all the cellular materials, including proteins, lipids and small functional compartments called organelles, that are required for normal cell activity. This proper apportioning of essential cell ingredients helps each cell get off to a running start.
This image shows an electron microscopy (EM) thin section taken at 10,000x magnification of a dividing yeast cell over-expressing the protein ubiquitin, which is involved in protein degradation and recycling. The picture features mother and daughter endosome accumulations (small organelles with internal vesicles), a darkly stained vacuole and a dividing nucleus in close contact with a cadre of lipid droplets (unstained spherical bodies). Other dynamic events are also visible, such as spindle microtubules in the nucleus and endocytic pits at the plasma membrane.
These extensive details were revealed thanks to a preservation method involving high-pressure freezing, freeze-substitution and Lowicryl HM20 embedding.
This image shows an electron microscopy (EM) thin section taken at 10,000x magnification of a dividing yeast cell over-expressing the protein ubiquitin, which is involved in protein degradation and recycling. The picture features mother and daughter endosome accumulations (small organelles with internal vesicles), a darkly stained vacuole and a dividing nucleus in close contact with a cadre of lipid droplets (unstained spherical bodies). Other dynamic events are also visible, such as spindle microtubules in the nucleus and endocytic pits at the plasma membrane.
These extensive details were revealed thanks to a preservation method involving high-pressure freezing, freeze-substitution and Lowicryl HM20 embedding.
Matthew West and Greg Odorizzi, University of Colorado
View Media
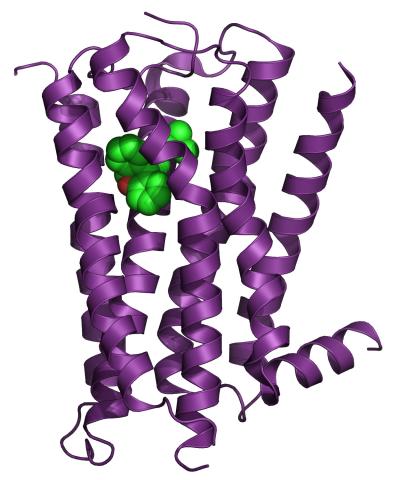
3360: H1 histamine receptor
3360: H1 histamine receptor
The receptor is shown bound to an inverse agonist, doxepin.
Raymond Stevens, The Scripps Research Institute
View Media
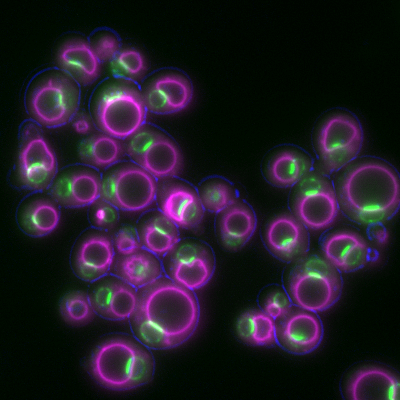
6772: Yeast cells responding to a glucose shortage
6772: Yeast cells responding to a glucose shortage
These yeast cells were exposed to a glucose (sugar) shortage. This caused the cells to compartmentalize HMGCR (green)—an enzyme involved in making cholesterol—to a patch on the nuclear envelope next to the vacuole/lysosome (purple). This process enhanced HMGCR activity and helped the yeast adapt to the glucose shortage. Researchers hope that understanding how yeast regulate cholesterol could ultimately lead to new ways to treat high cholesterol in people. This image was captured using a fluorescence microscope.
Mike Henne, University of Texas Southwestern Medical Center.
View Media
2802: Biosensors illustration
2802: Biosensors illustration
A rendering of an activity biosensor image overlaid with a cell-centered frame of reference used for image analysis of signal transduction. This is an example of NIH-supported research on single-cell analysis. Related to 2798 , 2799, 2800, 2801 and 2803.
Gaudenz Danuser, Harvard Medical School
View Media

1069: Lab mice
1069: Lab mice
Many researchers use the mouse (Mus musculus) as a model organism to study mammalian biology. Mice carry out practically all the same life processes as humans and, because of their small size and short generation times, are easily raised in labs. Scientists studying a certain cellular activity or disease can choose from tens of thousands of specially bred strains of mice to select those prone to developing certain tumors, neurological diseases, metabolic disorders, premature aging, or other conditions.
Bill Branson, National Institutes of Health
View Media
3616: Weblike sheath covering developing egg chambers in a giant grasshopper
3616: Weblike sheath covering developing egg chambers in a giant grasshopper
The lubber grasshopper, found throughout the southern United States, is frequently used in biology classes to teach students about the respiratory system of insects. Unlike mammals, which have red blood cells that carry oxygen throughout the body, insects have breathing tubes that carry air through their exoskeleton directly to where it's needed. This image shows the breathing tubes embedded in the weblike sheath cells that cover developing egg chambers.
This image was part of the Life: Magnified exhibit that ran from June 3, 2014, to January 21, 2015, at Dulles International Airport.
This image was part of the Life: Magnified exhibit that ran from June 3, 2014, to January 21, 2015, at Dulles International Airport.
Kevin Edwards, Johny Shajahan, and Doug Whitman, Illinois State University.
View Media
2304: Bacteria working to eat
2304: Bacteria working to eat
Gram-negative bacteria perform molecular acrobatics just to eat. Because they're encased by two membranes, they must haul nutrients across both. To test one theory of how the bacteria manage this feat, researchers used computer simulations of two proteins involved in importing vitamin B12. Here, the protein (red) anchored in the inner membrane of bacteria tugs on a much larger protein (green and blue) in the outer membrane. Part of the larger protein unwinds, creating a pore through which the vitamin can pass.
Emad Tajkhorshid, University of Illinois at Urbana-Champaign
View Media
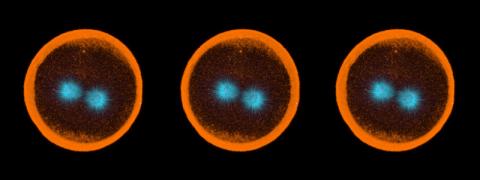
1050: Sea urchin embryo 04
1050: Sea urchin embryo 04
Stereo triplet of a sea urchin embryo stained to reveal actin filaments (orange) and microtubules (blue). This image is part of a series of images: image 1047, image 1048, image 1049, image 1051 and image 1052.
George von Dassow, University of Washington
View Media
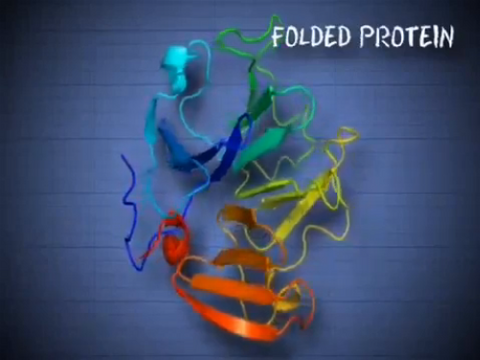
6602: See how immune cell acid destroys bacterial proteins
6602: See how immune cell acid destroys bacterial proteins
This animation shows the effect of exposure to hypochlorous acid, which is found in certain types of immune cells, on bacterial proteins. The proteins unfold and stick to one another, leading to cell death.
American Chemistry Council
View Media
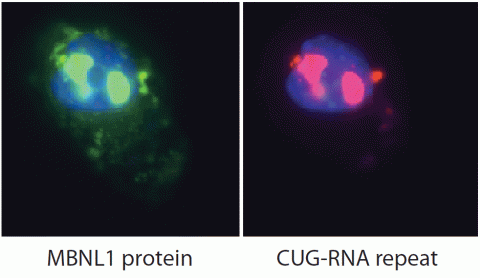
2727: Proteins related to myotonic dystrophy
2727: Proteins related to myotonic dystrophy
Myotonic dystrophy is thought to be caused by the binding of a protein called Mbnl1 to abnormal RNA repeats. In these two images of the same muscle precursor cell, the top image shows the location of the Mbnl1 splicing factor (green) and the bottom image shows the location of RNA repeats (red) inside the cell nucleus (blue). The white arrows point to two large foci in the cell nucleus where Mbnl1 is sequestered with RNA.
Manuel Ares, University of California, Santa Cruz
View Media
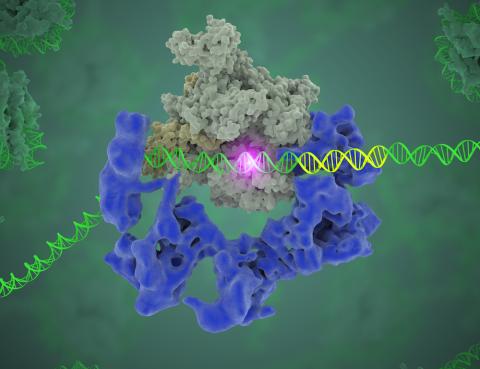
3766: TFIID complex binds DNA to start gene transcription
3766: TFIID complex binds DNA to start gene transcription
Gene transcription is a process by which the genetic information encoded in DNA is transcribed into RNA. It's essential for all life and requires the activity of proteins, called transcription factors, that detect where in a DNA strand transcription should start. In eukaryotes (i.e., those that have a nucleus and mitochondria), a protein complex comprising 14 different proteins is responsible for sniffing out transcription start sites and starting the process. This complex, called TFIID, represents the core machinery to which an enzyme, named RNA polymerase, can bind to and read the DNA and transcribe it to RNA. Scientists have used cryo-electron microscopy (cryo-EM) to visualize the TFIID-RNA polymerase-DNA complex in unprecedented detail. In this illustration, TFIID (blue) contacts the DNA and recruits the RNA polymerase (gray) for gene transcription. The start of the transcribed gene is shown with a flash of light. To learn more about the research that has shed new light on gene transcription, see this news release from Berkeley Lab. Related to video 5730.
Eva Nogales, Berkeley Lab
View Media
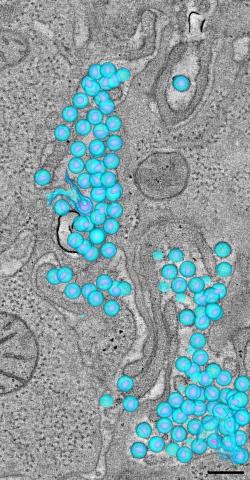
3571: HIV-1 virus in the colon
3571: HIV-1 virus in the colon
A tomographic reconstruction of the colon shows the location of large pools of HIV-1 virus particles (in blue) located in the spaces between adjacent cells. The purple objects within each sphere represent the conical cores that are one of the structural hallmarks of the HIV virus.
Mark Ladinsky, California Institute of Technology
View Media
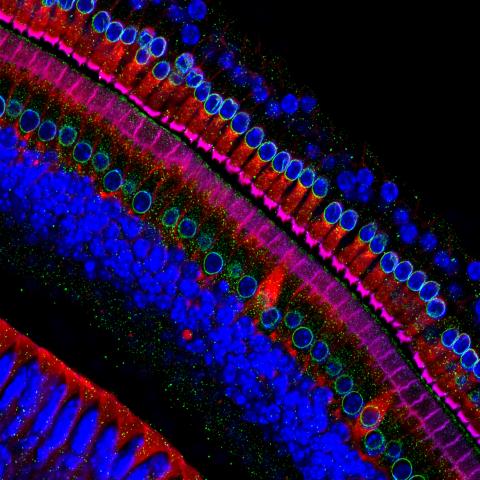
3618: Hair cells: the sound-sensing cells in the ear
3618: Hair cells: the sound-sensing cells in the ear
These cells get their name from the hairlike structures that extend from them into the fluid-filled tube of the inner ear. When sound reaches the ear, the hairs bend and the cells convert this movement into signals that are relayed to the brain. When we pump up the music in our cars or join tens of thousands of cheering fans at a football stadium, the noise can make the hairs bend so far that they actually break, resulting in long-term hearing loss.
This image was part of the Life: Magnified exhibit that ran from June 3, 2014, to January 21, 2015, at Dulles International Airport.
This image was part of the Life: Magnified exhibit that ran from June 3, 2014, to January 21, 2015, at Dulles International Airport.
Henning Horn, Brian Burke, and Colin Stewart, Institute of Medical Biology, Agency for Science, Technology, and Research, Singapore
View Media

1311: Housekeeping cell illustration
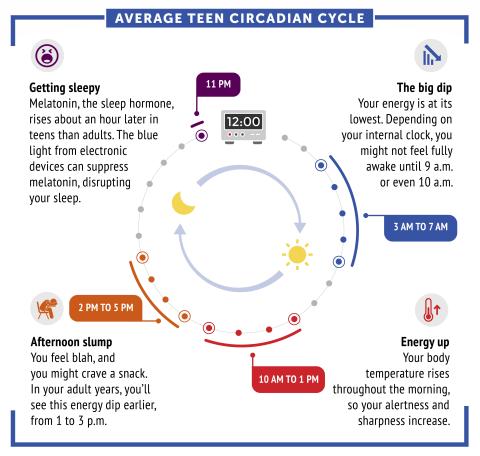
6611: Average teen circadian cycle
6611: Average teen circadian cycle
Circadian rhythms are physical, mental, and behavioral changes that follow a 24-hour cycle. Typical circadian rhythms lead to high energy during the middle of the day (10 a.m. to 1 p.m.) and an afternoon slump. At night, circadian rhythms cause the hormone melatonin to rise, making a person sleepy.
Learn more in NIGMS’ circadian rhythms featured topics page.
See 6612 for the Spanish version of this infographic.
Learn more in NIGMS’ circadian rhythms featured topics page.
See 6612 for the Spanish version of this infographic.
NIGMS
View Media
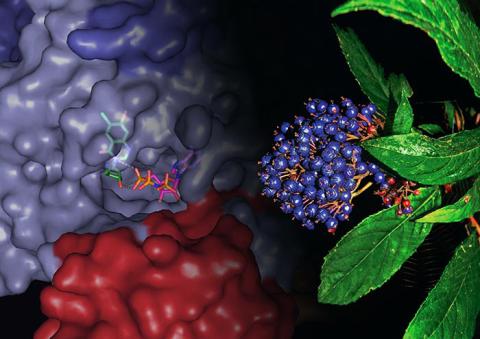
3483: Chang Shan
3483: Chang Shan
For thousands of years, Chinese herbalists have treated malaria using Chang Shan, a root extract from a type of hydrangea that grows in Tibet and Nepal. Recent studies have suggested Chang Shan can also reduce scar formation, treat multiple sclerosis and even slow cancer progression.
Paul Schimmel Lab, Scripps Research Institute
View Media
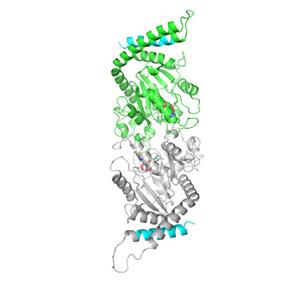
2744: Dynamin structure
2744: Dynamin structure
When a molecule arrives at a cell's outer membrane, the membrane creates a pouch around the molecule that protrudes inward. Directed by a protein called dynamin, the pouch then gets pinched off to form a vesicle that carries the molecule to the right place inside the cell. To better understand how dynamin performs its vital pouch-pinching role, researchers determined its structure. Based on the structure, they proposed that a dynamin "collar" at the pouch's base twists ever tighter until the vesicle pops free. Because cells absorb many drugs through vesicles, the discovery could lead to new drug delivery methods.
Josh Chappie, National Institute of Diabetes and Digestive and Kidney Diseases, NIH
View Media
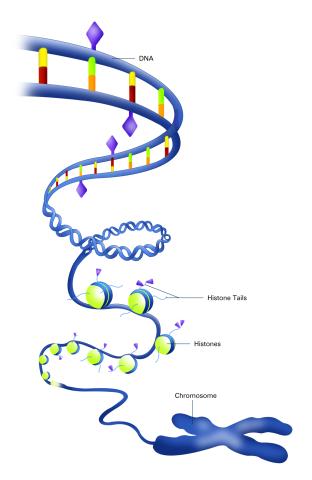
2563: Epigenetic code (with labels)
2563: Epigenetic code (with labels)
The "epigenetic code" controls gene activity with chemical tags that mark DNA (purple diamonds) and the "tails" of histone proteins (purple triangles). These markings help determine whether genes will be transcribed by RNA polymerase. Genes hidden from access to RNA polymerase are not expressed. See image 2562 for an unlabeled version of this illustration. Featured in The New Genetics.
Crabtree + Company
View Media
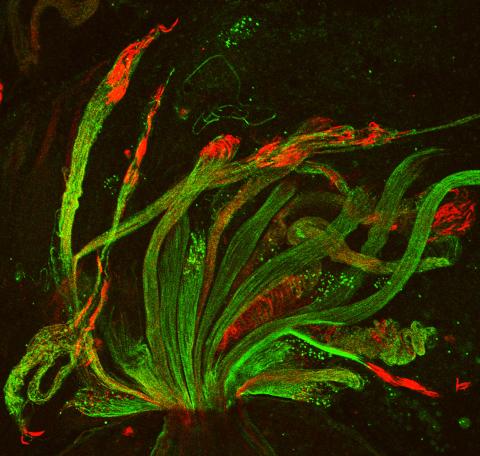
3590: Fruit fly spermatids
3590: Fruit fly spermatids
Developing spermatids (precursors of mature sperm cells) begin as small, round cells and mature into long-tailed, tadpole-shaped ones. In the sperm cell's head is the cell nucleus; in its tail is the power to outswim thousands of competitors to fertilize an egg. As seen in this microscopy image, fruit fly spermatids start out as groups of interconnected cells. A small lipid molecule called PIP2 helps spermatids tell their heads from their tails. Here, PIP2 (red) marks the nuclei and a cell skeleton-building protein called tubulin (green) marks the tails. When PIP2 levels are too low, some spermatids get mixed up and grow with their heads at the wrong end. Because sperm development is similar across species, studies in fruit flies could help researchers understand male infertility in humans.
Lacramioara Fabian, The Hospital for Sick Children, Toronto, Canada
View Media
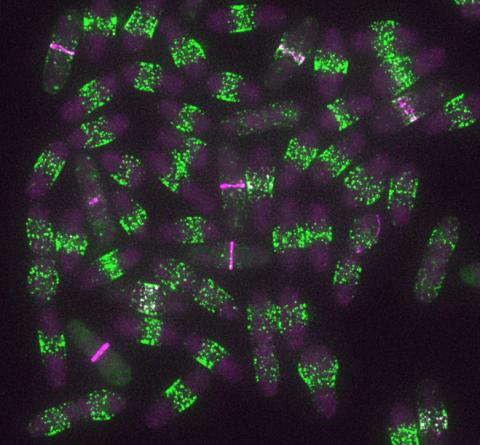
6791: Yeast cells entering mitosis
6791: Yeast cells entering mitosis
Yeast cells entering mitosis, also known as cell division. The green and magenta dots are two proteins that play important roles in mitosis. They show where the cells will split. This image was captured using wide-field microscopy with deconvolution.
Related to images 6792, 6793, 6794, 6797, 6798, and videos 6795 and 6796.
Related to images 6792, 6793, 6794, 6797, 6798, and videos 6795 and 6796.
Alaina Willet, Kathy Gould’s lab, Vanderbilt University.
View Media
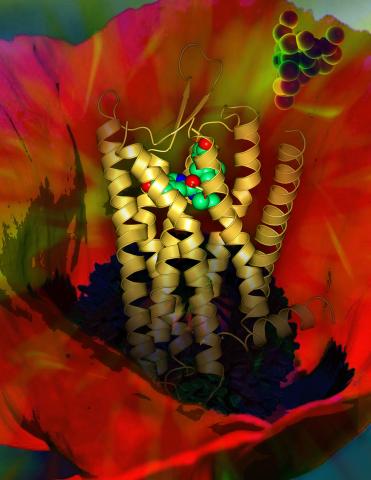
3314: Human opioid receptor structure superimposed on poppy
3314: Human opioid receptor structure superimposed on poppy
Opioid receptors on the surfaces of brain cells are involved in pleasure, pain, addiction, depression, psychosis, and other conditions. The receptors bind to both innate opioids and drugs ranging from hospital anesthetics to opium. Researchers at The Scripps Research Institute, supported by the NIGMS Protein Structure Initiative, determined the first three-dimensional structure of a human opioid receptor, a kappa-opioid receptor. In this illustration, the submicroscopic receptor structure is shown while bound to an agonist (or activator). The structure is superimposed on a poppy flower, the source of opium.
Raymond Stevens, The Scripps Research Institute
View Media
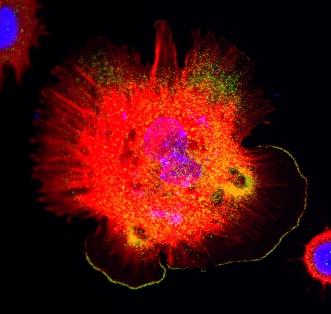
3328: Spreading Cells 01
3328: Spreading Cells 01
Cells move forward with lamellipodia and filopodia supported by networks and bundles of actin filaments. Proper, controlled cell movement is a complex process. Recent research has shown that an actin-polymerizing factor called the Arp2/3 complex is the key component of the actin polymerization engine that drives amoeboid cell motility. ARPC3, a component of the Arp2/3 complex, plays a critical role in actin nucleation. In this photo, the ARPC3+/+ fibroblast cells were fixed and stained with Alexa 546 phalloidin for F-actin (red), Arp2 (green), and DAPI to visualize the nucleus (blue). Arp2, a subunit of the Arp2/3 complex, is localized at the lamellipodia leading edge of ARPC3+/+ fibroblast cells. Related to images 3329, 3330, 3331, 3332, and 3333.
Rong Li and Praveen Suraneni, Stowers Institute for Medical Research
View Media
