Switch to List View
Image and Video Gallery
This is a searchable collection of scientific photos, illustrations, and videos. The images and videos in this gallery are licensed under Creative Commons Attribution Non-Commercial ShareAlike 3.0. This license lets you remix, tweak, and build upon this work non-commercially, as long as you credit and license your new creations under identical terms.
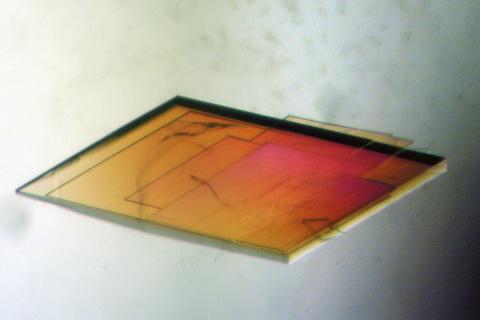
2403: Pig trypsin crystal
2403: Pig trypsin crystal
A crystal of pig trypsin protein created for X-ray crystallography, which can reveal detailed, three-dimensional protein structures.
Alex McPherson, University of California, Irvine
View Media
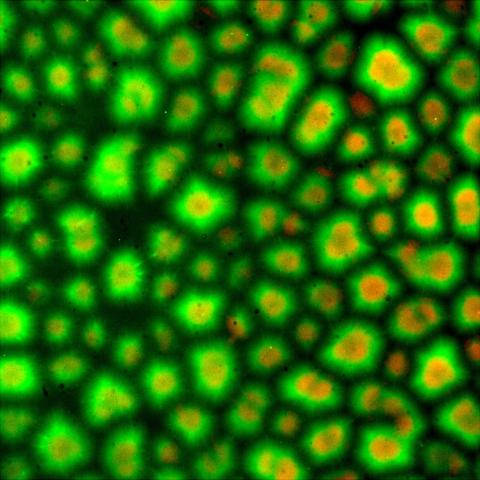
6586: Cell-like compartments from frog eggs 3
6586: Cell-like compartments from frog eggs 3
Cell-like compartments that spontaneously emerged from scrambled frog eggs. Endoplasmic reticulum (red) and microtubules (green) are visible. Image created using epifluorescence microscopy.
For more photos of cell-like compartments from frog eggs view: 6584, 6585, 6591, 6592, and 6593.
For videos of cell-like compartments from frog eggs view: 6587, 6588, 6589, and 6590.
Xianrui Cheng, Stanford University School of Medicine.
View Media
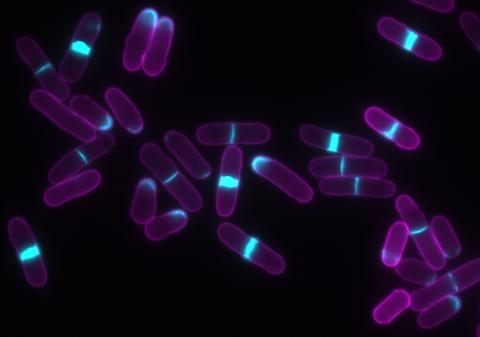
6797: Yeast cells with accumulated cell wall material
6797: Yeast cells with accumulated cell wall material
Yeast cells that abnormally accumulate cell wall material (blue) at their ends and, when preparing to divide, in their middles. This image was captured using wide-field microscopy with deconvolution.
Related to images 6791, 6792, 6793, 6794, 6798, and videos 6795 and 6796.
Related to images 6791, 6792, 6793, 6794, 6798, and videos 6795 and 6796.
Alaina Willet, Kathy Gould’s lab, Vanderbilt University.
View Media
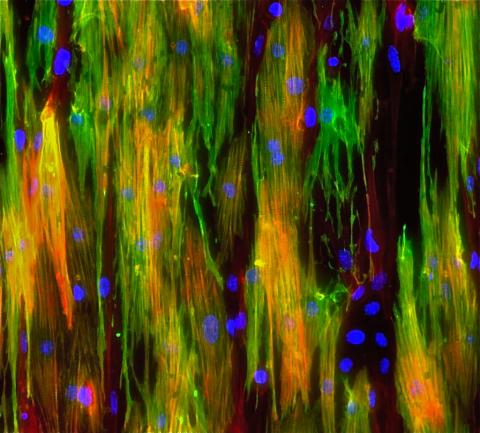
3283: Mouse heart muscle cells 02
3283: Mouse heart muscle cells 02
This image shows neonatal mouse heart cells. These cells were grown in the lab on a chip that aligns the cells in a way that mimics what is normally seen in the body. Green shows the muscle protein toponin I. Red indicates the muscle protein actin, and blue indicates the cell nuclei. The work shown here was part of a study attempting to grow heart tissue in the lab to repair damage after a heart attack. Image and caption information courtesy of the California Institute for Regenerative Medicine. Related to images 3281 and 3282.
Kara McCloskey lab, University of California, Merced, via CIRM
View Media
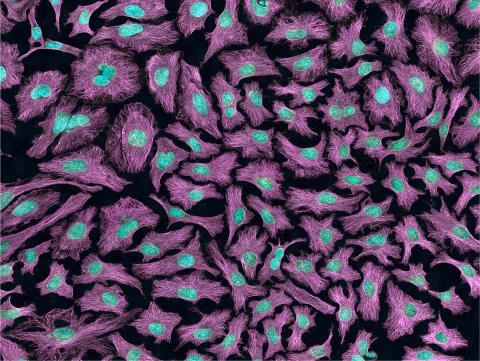
3520: HeLa cells
3520: HeLa cells
Multiphoton fluorescence image of HeLa cells with cytoskeletal microtubules (magenta) and DNA (cyan). Nikon RTS2000MP custom laser scanning microscope. See related images 3518, 3519, 3521, 3522.
National Center for Microscopy and Imaging Research (NCMIR)
View Media
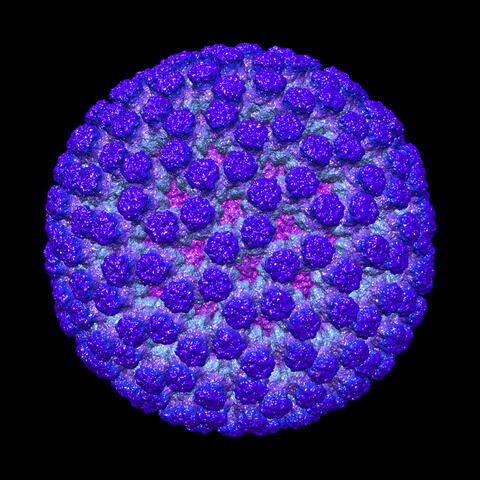
3584: Rotavirus structure
3584: Rotavirus structure
This image shows a computer-generated, three-dimensional map of the rotavirus structure. This virus infects humans and other animals and causes severe diarrhea in infants and young children. By the age of five, almost every child in the world has been infected with this virus at least once. Scientists have found a vaccine against rotavirus, so in the United States there are very few fatalities, but in developing countries and in places where the vaccine is unavailable, this virus is responsible for more than 200,000 deaths each year.
The rotavirus comprises three layers: the outer, middle and inner layers. On infection, the outer layer is removed, leaving behind a "double-layered particle." Researchers have studied the structure of this double-layered particle with a transmission electron microscope. Many images of the virus at a magnification of ~50,000x were acquired, and computational analysis was used to combine the individual particle images into a three-dimensional reconstruction.
The image was rendered by Melody Campbell (PhD student at TSRI). Work that led to the 3D map was published in Campbell et al. Movies of ice-embedded particles enhance resolution in electron cryo-microscopy. Structure. 2012;20(11):1823-8. PMCID: PMC3510009.
This image was part of the Life: Magnified exhibit that ran from June 3, 2014, to January 21, 2015, at Dulles International Airport.
The rotavirus comprises three layers: the outer, middle and inner layers. On infection, the outer layer is removed, leaving behind a "double-layered particle." Researchers have studied the structure of this double-layered particle with a transmission electron microscope. Many images of the virus at a magnification of ~50,000x were acquired, and computational analysis was used to combine the individual particle images into a three-dimensional reconstruction.
The image was rendered by Melody Campbell (PhD student at TSRI). Work that led to the 3D map was published in Campbell et al. Movies of ice-embedded particles enhance resolution in electron cryo-microscopy. Structure. 2012;20(11):1823-8. PMCID: PMC3510009.
This image was part of the Life: Magnified exhibit that ran from June 3, 2014, to January 21, 2015, at Dulles International Airport.
Bridget Carragher, The Scripps Research Institute, La Jolla, CA
View Media
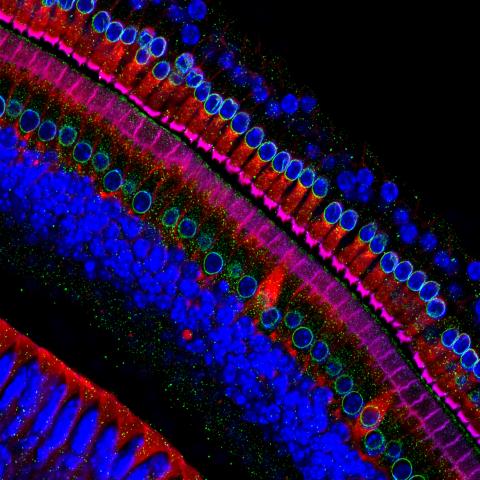
3618: Hair cells: the sound-sensing cells in the ear
3618: Hair cells: the sound-sensing cells in the ear
These cells get their name from the hairlike structures that extend from them into the fluid-filled tube of the inner ear. When sound reaches the ear, the hairs bend and the cells convert this movement into signals that are relayed to the brain. When we pump up the music in our cars or join tens of thousands of cheering fans at a football stadium, the noise can make the hairs bend so far that they actually break, resulting in long-term hearing loss.
This image was part of the Life: Magnified exhibit that ran from June 3, 2014, to January 21, 2015, at Dulles International Airport.
This image was part of the Life: Magnified exhibit that ran from June 3, 2014, to January 21, 2015, at Dulles International Airport.
Henning Horn, Brian Burke, and Colin Stewart, Institute of Medical Biology, Agency for Science, Technology, and Research, Singapore
View Media

3592: Math from the heart
3592: Math from the heart
Watch a cell ripple toward a beam of light that turns on a movement-related protein.
View Media
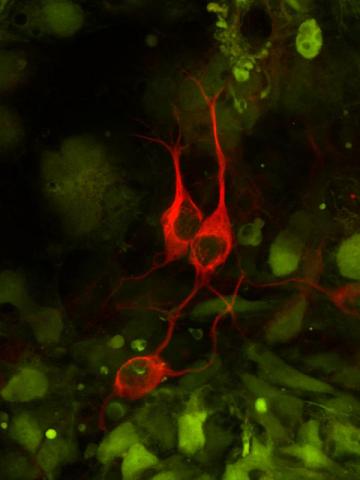
3290: Three neurons and human ES cells
3290: Three neurons and human ES cells
The three neurons (red) visible in this image were derived from human embryonic stem cells. Undifferentiated stem cells are green here. Image and caption information courtesy of the California Institute for Regenerative Medicine.
Anirvan Ghosh lab, University of California, San Diego, via CIRM
View Media
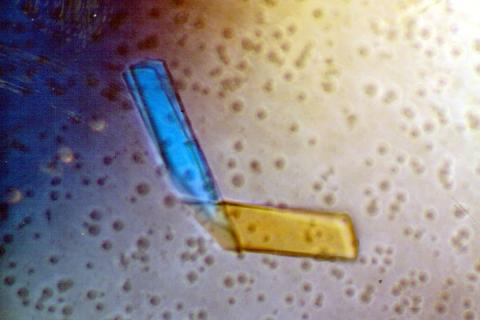
2399: Bence Jones protein MLE
2399: Bence Jones protein MLE
A crystal of Bence Jones protein created for X-ray crystallography, which can reveal detailed, three-dimensional protein structures.
Alex McPherson, University of California, Irvine
View Media
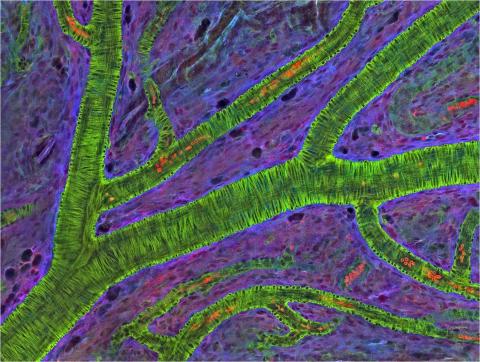
3400: Small blood vessels in a mouse retina
3400: Small blood vessels in a mouse retina
Blood vessels at the back of the eye (retina) are used to diagnose glaucoma and diabetic eye disease. They also display characteristic changes in people with high blood pressure. In the image, the vessels appear green. It's not actually the vessels that are stained green, but rather filaments of a protein called actin that wraps around the vessels. Most of the red blood cells were replaced by fluid as the tissue was prepared for the microscope. The tiny red dots are red blood cells that remain in the vessels. The image was captured using confocal and 2-photon excitation microscopy for a project related to neurofibromatosis.
National Center for Microscopy and Imaging Research
View Media
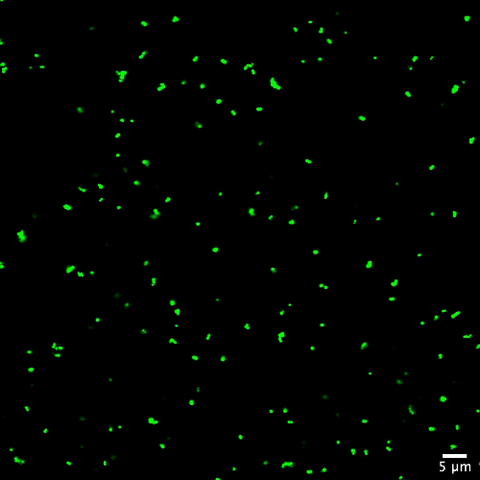
6805: Staphylococcus aureus aggregating upon contact with synovial fluid
6805: Staphylococcus aureus aggregating upon contact with synovial fluid
Staphylococcus aureus bacteria (green) grouping together upon contact with synovial fluid—a viscous substance found in joints. The formation of groups can help protect the bacteria from immune system defenses and from antibiotics, increasing the likelihood of an infection. This video is a 1-hour time lapse and was captured using a confocal laser scanning microscope.
More information about the research that produced this video can be found in the Journal of Bacteriology paper "In Vitro Staphylococcal Aggregate Morphology and Protection from Antibiotics Are Dependent on Distinct Mechanisms Arising from Postsurgical Joint Components and Fluid Motion" by Staats et al.
Related to images 6803 and 6804.
More information about the research that produced this video can be found in the Journal of Bacteriology paper "In Vitro Staphylococcal Aggregate Morphology and Protection from Antibiotics Are Dependent on Distinct Mechanisms Arising from Postsurgical Joint Components and Fluid Motion" by Staats et al.
Related to images 6803 and 6804.
Paul Stoodley, The Ohio State University.
View Media
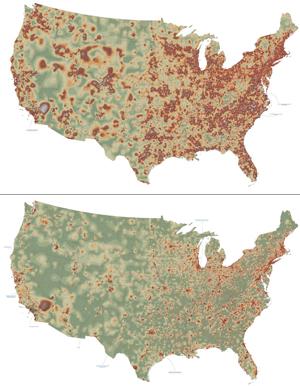
2320: Mapping disease spread
2320: Mapping disease spread
How far and fast an infectious disease spreads across a community depends on many factors, including transportation. These U.S. maps, developed as part of an international study to simulate and analyze disease spread, chart daily commuting patterns. They show where commuters live (top) and where they travel for work (bottom). Green represents the fewest number of people whereas orange, brown, and white depict the most. Such information enables researchers and policymakers to visualize how an outbreak in one area can spread quickly across a geographic region.
David Chrest, RTI International
View Media
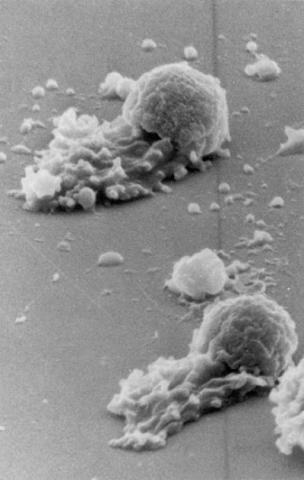
3489: Worm sperm
3489: Worm sperm
To develop a system for studying cell motility in unnatrual conditions -- a microscope slide instead of the body -- Tom Roberts and Katsuya Shimabukuro at Florida State University disassembled and reconstituted the motility parts used by worm sperm cells.
Tom Roberts, Florida State University
View Media
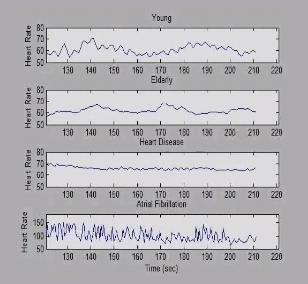
3596: Heart rates time series image
3596: Heart rates time series image
These time series show the heart rates of four different individuals. Automakers use steel scraps to build cars, construction companies repurpose tires to lay running tracks, and now scientists are reusing previously discarded medical data to better understand our complex physiology. Through a website called PhysioNet developed in part by Beth Israel Deaconess Medical Center cardiologist Ary Goldberger, scientists can access complete physiologic recordings, such as heart rate, respiration, brain activity and gait. They then can use free software to analyze the data and find patterns in it. The patterns could ultimately help health care professionals diagnose and treat health conditions like congestive heart failure, sleeping disorders, epilepsy and walking problems. PhysioNet is supported by NIH's National Institute of Biomedical Imaging and Bioengineering as well as by NIGMS.
Madalena Costa and Ary Goldberger, Beth Israel Deaconess Medical Center
View Media
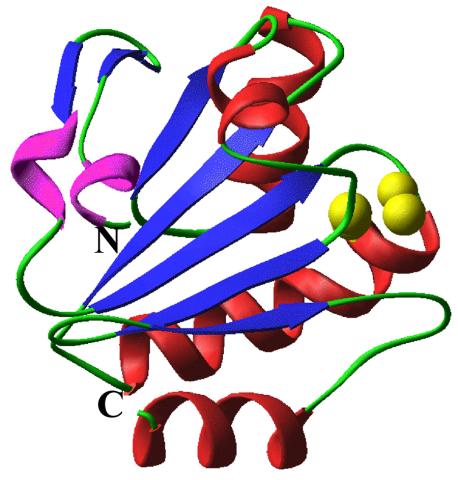
2382: PanB from M. tuberculosis (2)
2382: PanB from M. tuberculosis (2)
Model of an enzyme, PanB, from Mycobacterium tuberculosis, the bacterium that causes most cases of tuberculosis. This enzyme is an attractive drug target.
Mycobacterium Tuberculosis Center, PSI-1
View Media
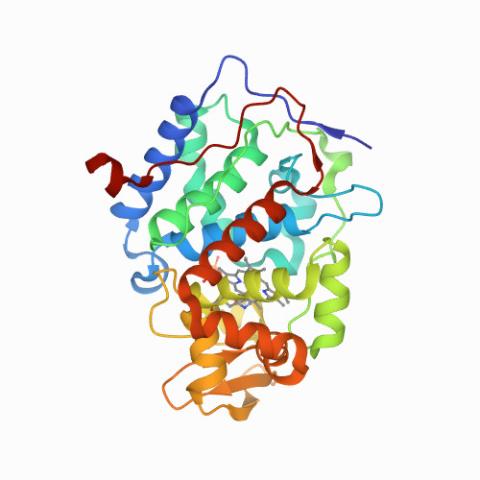
6762: CCP enzyme
6762: CCP enzyme
The enzyme CCP is found in the mitochondria of baker’s yeast. Scientists study the chemical reactions that CCP triggers, which involve a water molecule, iron, and oxygen. This structure was determined using an X-ray free electron laser.
Protein Data Bank.
View Media
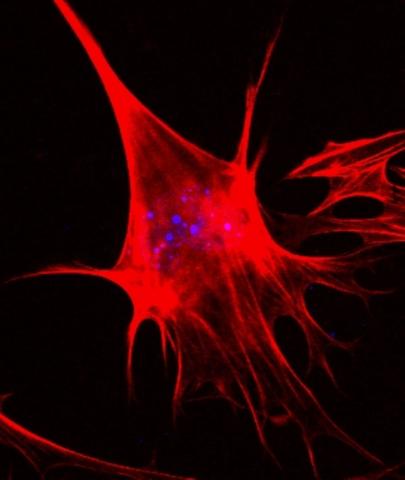
3333: Polarized cells- 02
3333: Polarized cells- 02
Cells move forward with lamellipodia and filopodia supported by networks and bundles of actin filaments. Proper, controlled cell movement is a complex process. Recent research has shown that an actin-polymerizing factor called the Arp2/3 complex is the key component of the actin polymerization engine that drives amoeboid cell motility. ARPC3, a component of the Arp2/3 complex, plays a critical role in actin nucleation. In this photo, the ARPC3-/- fibroblast cells were fixed and stained with Alexa 546 phalloidin for F-actin (red) and DAPI to visualize the nucleus (blue). In the absence of functional Arp2/3 complex, ARPC3-/- fibroblast cells' leading edge morphology is significantly altered with filopodia-like structures. Related to images 3328, 3329, 3330, 3331, and 3332.
Rong Li and Praveen Suraneni, Stowers Institute for Medical Research
View Media
1293: Sperm cell
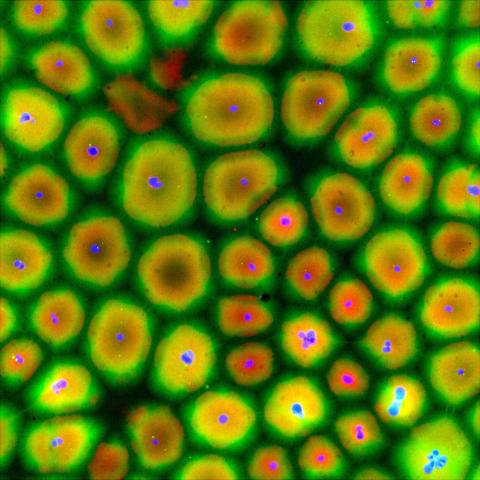
6584: Cell-like compartments from frog eggs
6584: Cell-like compartments from frog eggs
Cell-like compartments that spontaneously emerged from scrambled frog eggs, with nuclei (blue) from frog sperm. Endoplasmic reticulum (red) and microtubules (green) are also visible. Image created using epifluorescence microscopy.
For more photos of cell-like compartments from frog eggs view: 6585, 6586, 6591, 6592, and 6593.
For videos of cell-like compartments from frog eggs view: 6587, 6588, 6589, and 6590.
Xianrui Cheng, Stanford University School of Medicine.
View Media
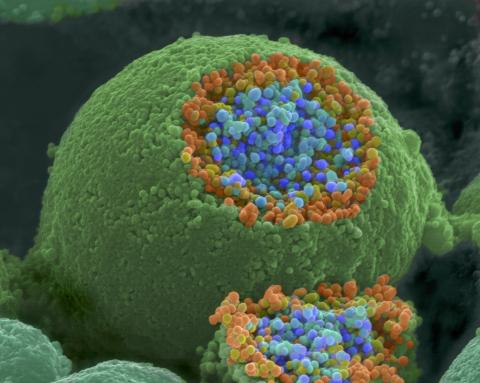
1244: Nerve ending
1244: Nerve ending
A scanning electron microscope picture of a nerve ending. It has been broken open to reveal vesicles (orange and blue) containing chemicals used to pass messages in the nervous system.
Tina Weatherby Carvalho, University of Hawaii at Manoa
View Media
1337: Bicycling cell
1337: Bicycling cell
A humorous treatment of the concept of a cycling cell.
Judith Stoffer
View Media
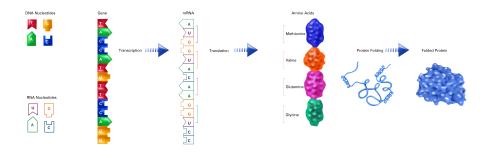
2510: From DNA to Protein (labeled)
2510: From DNA to Protein (labeled)
The genetic code in DNA is transcribed into RNA, which is translated into proteins with specific sequences. During transcription, nucleotides in DNA are copied into RNA, where they are read three at a time to encode the amino acids in a protein. Many parts of a protein fold as the amino acids are strung together.
See image 2509 for an unlabeled version of this illustration.
Featured in The Structures of Life.
See image 2509 for an unlabeled version of this illustration.
Featured in The Structures of Life.
Crabtree + Company
View Media

2305: Beaded bacteriophage
2305: Beaded bacteriophage
This sculpture made of purple and clear glass beads depicts bacteriophage Phi174, a virus that infects bacteria. It rests on a surface that portrays an adaptive landscape, a conceptual visualization. The ridges represent the gene combinations associated with the greatest fitness levels of the virus, as measured by how quickly the virus can reproduce itself. Phi174 is an important model system for studies of viral evolution because its genome can readily be sequenced as it evolves under defined laboratory conditions.
Holly Wichman, University of Idaho. (Surface by A. Johnston; photo by J. Palmersheim)
View Media
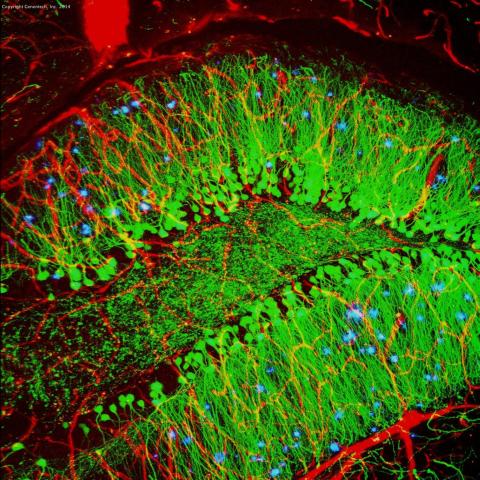
3604: Brain showing hallmarks of Alzheimer's disease
3604: Brain showing hallmarks of Alzheimer's disease
Along with blood vessels (red) and nerve cells (green), this mouse brain shows abnormal protein clumps known as plaques (blue). These plaques multiply in the brains of people with Alzheimer's disease and are associated with the memory impairment characteristic of the disease. Because mice have genomes nearly identical to our own, they are used to study both the genetic and environmental factors that trigger Alzheimer's disease. Experimental treatments are also tested in mice to identify the best potential therapies for human patients.
This image was part of the Life: Magnified exhibit that ran from June 3, 2014, to January 21, 2015, at Dulles International Airport.
This image was part of the Life: Magnified exhibit that ran from June 3, 2014, to January 21, 2015, at Dulles International Airport.
Alvin Gogineni, Genentech
View Media
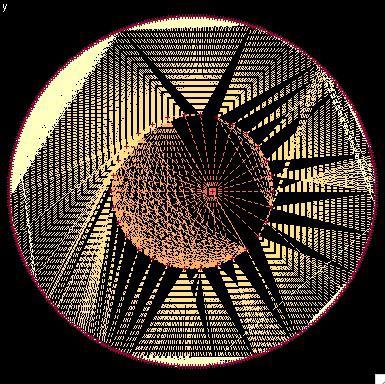
2322: Modeling disease spread
2322: Modeling disease spread
What looks like a Native American dream catcher is really a network of social interactions within a community. The red dots along the inner and outer circles represent people, while the different colored lines represent direct contact between them. All connections originate from four individuals near the center of the graph. Modeling social networks can help researchers understand how diseases spread.
Stephen Eubank, University of Virginia Biocomplexity Institute (formerly Virginia Bioinformatics Institute)
View Media
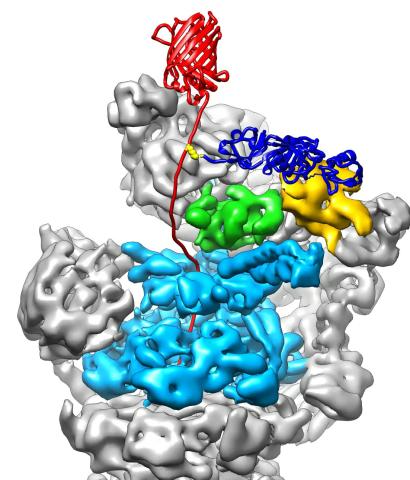
3763: The 26S proteasome engages with a protein substrate
3763: The 26S proteasome engages with a protein substrate
The proteasome is a critical multiprotein complex in the cell that breaks down and recycles proteins that have become damaged or are no longer needed. This illustration shows a protein substrate (red) that is bound through its ubiquitin chain (blue) to one of the ubiquitin receptors of the proteasome (Rpn10, yellow). The substrate's flexible engagement region gets engaged by the AAA+ motor of the proteasome (cyan), which initiates mechanical pulling, unfolding and movement of the protein into the proteasome's interior for cleavage into small shorter protein pieces called peptides. During movement of the substrate, its ubiquitin modification gets cleaved off by the deubiquitinase Rpn11 (green), which sits directly above the entrance to the AAA+ motor pore and acts as a gatekeeper to ensure efficient ubiquitin removal, a prerequisite for fast protein breakdown by the 26S proteasome. Related to video 3764.
Andreas Martin, HHMI
View Media
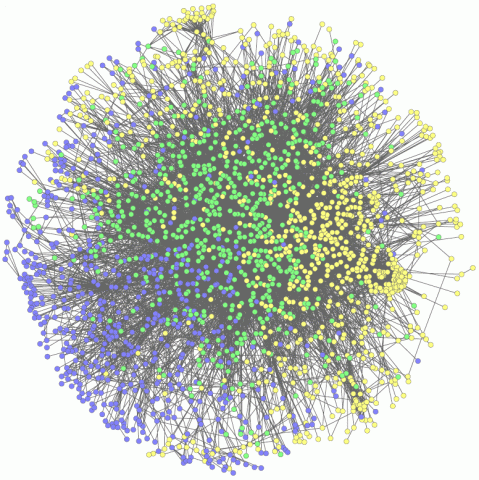
2735: Network Map
2735: Network Map
This network map shows the overlap (green) between the long QT syndrome (yellow) and epilepsy (blue) protein-interaction neighborhoods located within the human interactome. Researchers have learned to integrate genetic, cellular and clinical information to find out why certain medicines can trigger fatal heart arrhythmias. Featured in Computing Life magazine.
Seth Berger, Mount Sinai School of Medicine
View Media
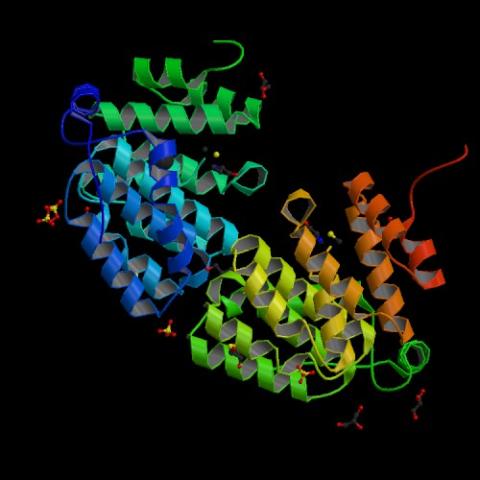
2342: Protein from E. faecalis
2342: Protein from E. faecalis
X-ray structure of a DNA repair enzyme superfamily representative from the human gastrointestinal bacterium Enterococcus faecalis. European scientists used this structure to generate homologous structures. Featured as the May 2007 Protein Structure Initiative Structure of the Month.
Midwest Center for Structural Genomics
View Media

1275: Golgi
1275: Golgi
The Golgi complex, also called the Golgi apparatus or, simply, the Golgi. This organelle receives newly made proteins and lipids from the ER, puts the finishing touches on them, addresses them, and sends them to their final destinations.
Judith Stoffer
View Media
2752: Bacterial spore
2752: Bacterial spore
A spore from the bacterium Bacillus subtilis shows four outer layers that protect the cell from harsh environmental conditions.
Patrick Eichenberger, New York University
View Media
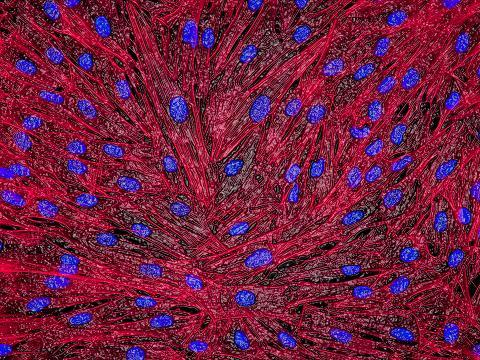
3670: DNA and actin in cultured fibroblast cells
3670: DNA and actin in cultured fibroblast cells
DNA (blue) and actin (red) in cultured fibroblast cells.
Tom Deerinck, National Center for Microscopy and Imaging Research (NCMIR)
View Media
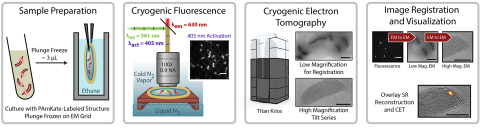
6568: Correlative imaging by annotation with single molecules (CIASM) process
6568: Correlative imaging by annotation with single molecules (CIASM) process
These images illustrate a technique combining cryo-electron tomography and super-resolution fluorescence microscopy called correlative imaging by annotation with single molecules (CIASM). CIASM enables researchers to identify small structures and individual molecules in cells that they couldn’t using older techniques.
Peter Dahlberg, Stanford University.
View Media
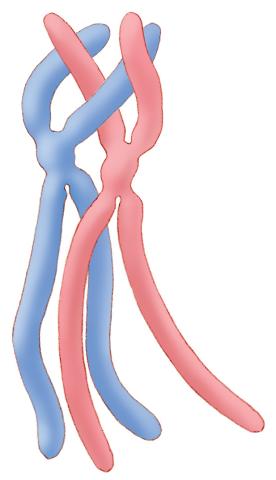
1315: Chromosomes before crossing over
1315: Chromosomes before crossing over
Duplicated pair of chromosomes lined up and ready to cross over.
Judith Stoffer
View Media
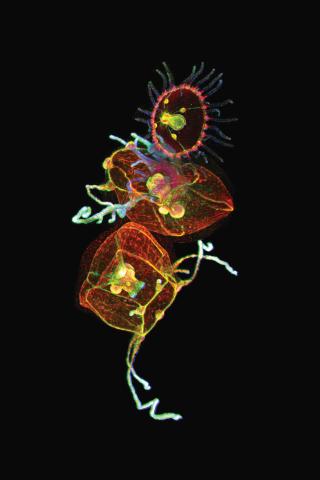
3636: Jellyfish, viewed with ZEISS Lightsheet Z.1 microscope
3636: Jellyfish, viewed with ZEISS Lightsheet Z.1 microscope
Jellyfish are especially good models for studying the evolution of embryonic tissue layers. Despite being primitive, jellyfish have a nervous system (stained green here) and musculature (red). Cell nuclei are stained blue. By studying how tissues are distributed in this simple organism, scientists can learn about the evolution of the shapes and features of diverse animals.
This image was part of the Life: Magnified exhibit that ran from June 3, 2014, to January 21, 2015, at Dulles International Airport.
This image was part of the Life: Magnified exhibit that ran from June 3, 2014, to January 21, 2015, at Dulles International Airport.
Helena Parra, Pompeu Fabra University, Spain
View Media
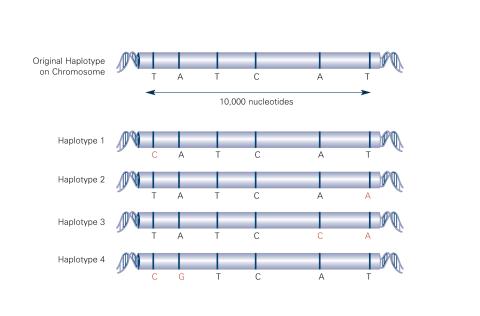
2567: Haplotypes (with labels)
2567: Haplotypes (with labels)
Haplotypes are combinations of gene variants that are likely to be inherited together within the same chromosomal region. In this example, an original haplotype (top) evolved over time to create three newer haplotypes that each differ by a few nucleotides (red). See image 2566 for an unlabeled version of this illustration. Featured in The New Genetics.
Crabtree + Company
View Media
1084: Natcher Building 04
1084: Natcher Building 04
NIGMS staff are located in the Natcher Building on the NIH campus.
Alisa Machalek, National Institute of General Medical Sciences
View Media
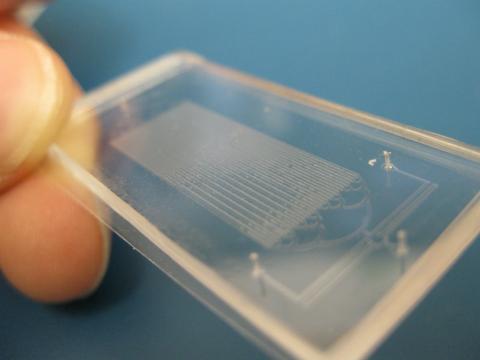
3265: Microfluidic chip
3265: Microfluidic chip
Microfluidic chips have many uses in biology labs. The one shown here was used by bioengineers to study bacteria, allowing the researchers to synchronize their fluorescing so they would blink in unison. Related to images 3266 and 3268. From a UC San Diego news release, "Researchers create living 'neon signs' composed of millions of glowing bacteria."
Jeff Hasty Lab, UC San Diego
View Media

2593: Precise development in the fruit fly embryo
2593: Precise development in the fruit fly embryo
This 2-hour-old fly embryo already has a blueprint for its formation, and the process for following it is so precise that the difference of just a few key molecules can change the plans. Here, blue marks a high concentration of Bicoid, a key signaling protein that directs the formation of the fly's head. It also regulates another important protein, Hunchback (green), that further maps the head and thorax structures and partitions the embryo in half (red is DNA). The yellow dots overlaying the embryo plot the concentration of Bicoid versus Hunchback proteins within each nucleus. The image illustrates the precision with which an embryo interprets and locates its halfway boundary, approaching limits set by simple physical principles. This image was a finalist in the 2008 Drosophila Image Award.
Thomas Gregor, Princeton University
View Media

2576: Cone snail shell
2576: Cone snail shell
A shell from the venomous cone snail Conus omaria, which lives in the Pacific and Indian oceans and eats other snails. University of Utah scientists discovered a new toxin in this snail species' venom, and say it will be a useful tool in designing new medicines for a variety of brain disorders, including Alzheimer's and Parkinson's diseases, depression, nicotine addiction and perhaps schizophrenia.
Kerry Matz, University of Utah
View Media

6803: Staphylococcus aureus aggregates on microstructured titanium surface
6803: Staphylococcus aureus aggregates on microstructured titanium surface
Groups of Staphylococcus aureus bacteria (blue) attached to a microstructured titanium surface (green) that mimics an orthopedic implant used in joint replacement. The attachment of pre-formed groups of bacteria may lead to infections because the groups can tolerate antibiotics and evade the immune system. This image was captured using a scanning electron microscope.
More information on the research that produced this image can be found in the Antibiotics paper "Free-floating aggregate and single-cell-initiated biofilms of Staphylococcus aureus" by Gupta et al.
Related to image 6804 and video 6805.
More information on the research that produced this image can be found in the Antibiotics paper "Free-floating aggregate and single-cell-initiated biofilms of Staphylococcus aureus" by Gupta et al.
Related to image 6804 and video 6805.
Paul Stoodley, The Ohio State University.
View Media
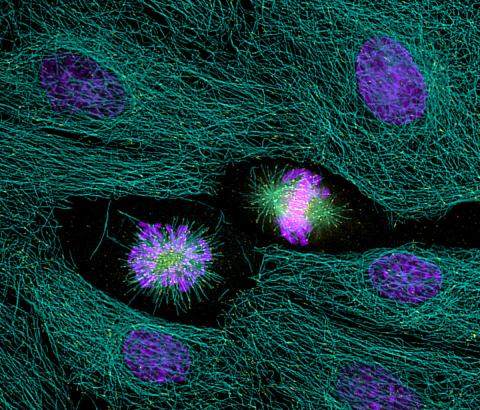
2429: Highlighted cells
2429: Highlighted cells
The cytoskeleton (green) and DNA (purple) are highlighed in these cells by immunofluorescence.
Torsten Wittmann, Scripps Research Institute
View Media

6550: Time-lapse video of floral pattern in a mixture of two bacterial species, Acinetobacter baylyi and Escherichia coli, grown on a semi-solid agar for 24 hours
6550: Time-lapse video of floral pattern in a mixture of two bacterial species, Acinetobacter baylyi and Escherichia coli, grown on a semi-solid agar for 24 hours
This time-lapse video shows the emergence of a flower-like pattern in a mixture of two bacterial species, motile Acinetobacter baylyi and non-motile Escherichia coli (green), that are grown together for 24 hours on 0.75% agar surface from a small inoculum in the center of a Petri dish.
See 6557 for a photo of this process at 24 hours on 0.75% agar surface.
See 6553 for a photo of this process at 48 hours on 1% agar surface.
See 6555 for another photo of this process at 48 hours on 1% agar surface.
See 6556 for a photo of this process at 72 hours on 0.5% agar surface.
See 6557 for a photo of this process at 24 hours on 0.75% agar surface.
See 6553 for a photo of this process at 48 hours on 1% agar surface.
See 6555 for another photo of this process at 48 hours on 1% agar surface.
See 6556 for a photo of this process at 72 hours on 0.5% agar surface.
L. Xiong et al, eLife 2020;9: e48885
View Media
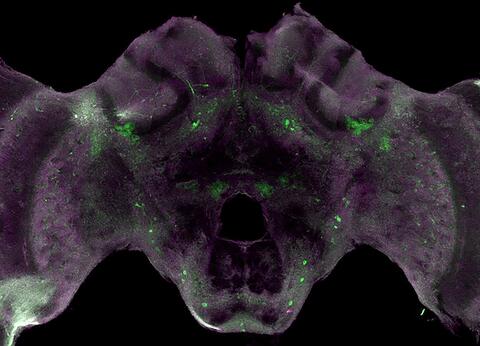
6755: Honeybee brain
6755: Honeybee brain
Insect brains, like the honeybee brain shown here, are very different in shape from human brains. Despite that, bee and human brains have a lot in common, including many of the genes and neurochemicals they rely on in order to function. The bright-green spots in this image indicate the presence of tyrosine hydroxylase, an enzyme that allows the brain to produce dopamine. Dopamine is involved in many important functions, such as the ability to experience pleasure. This image was captured using confocal microscopy.
Gene Robinson, University of Illinois at Urbana-Champaign.
View Media
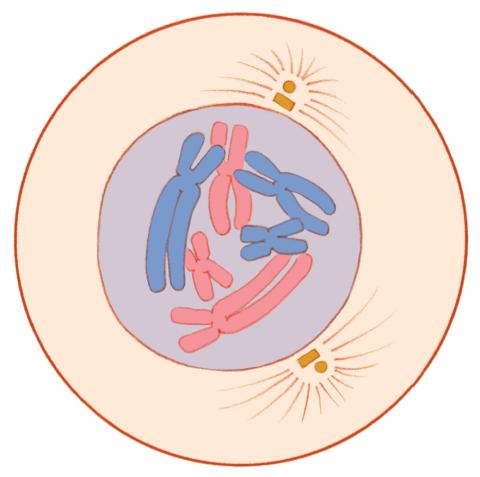
1330: Mitosis - prophase
1330: Mitosis - prophase
A cell in prophase, near the start of mitosis: In the nucleus, chromosomes condense and become visible. In the cytoplasm, the spindle forms. Mitosis is responsible for growth and development, as well as for replacing injured or worn out cells throughout the body. For simplicity, mitosis is illustrated here with only six chromosomes.
Judith Stoffer
View Media
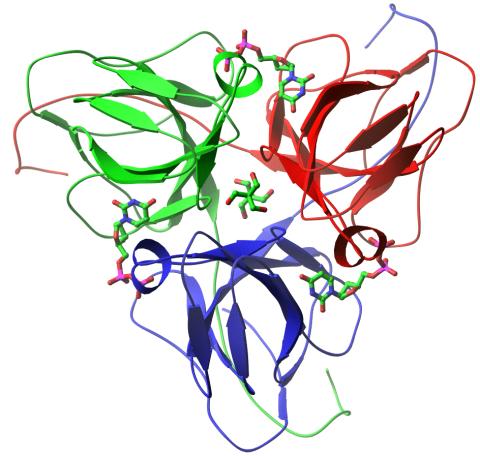
2381: dUTP pyrophosphatase from M. tuberculosis
2381: dUTP pyrophosphatase from M. tuberculosis
Model of an enzyme, dUTP pyrophosphatase, from Mycobacterium tuberculosis. Drugs targeted to this enzyme might inhibit the replication of the bacterium that causes most cases of tuberculosis.
Mycobacterium Tuberculosis Center, PSI
View Media
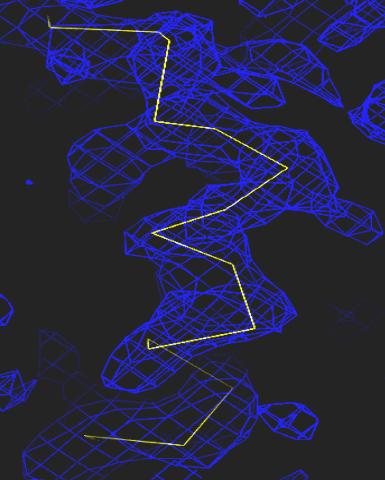
2354: Section of an electron density map
2354: Section of an electron density map
Electron density maps such as this one are generated from the diffraction patterns of X-rays passing through protein crystals. These maps are then used to generate a model of the protein's structure by fitting the protein's amino acid sequence (yellow) into the observed electron density (blue).
The Southeast Collaboratory for Structural Genomics
View Media
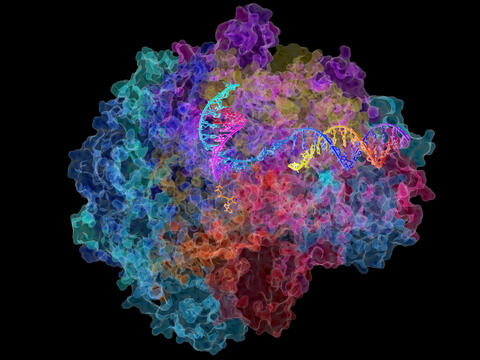
2484: RNA Polymerase II
2484: RNA Polymerase II
NIGMS-funded researchers led by Roger Kornberg solved the structure of RNA polymerase II. This is the enzyme in mammalian cells that catalyzes the transcription of DNA into messenger RNA, the molecule that in turn dictates the order of amino acids in proteins. For his work on the mechanisms of mammalian transcription, Kornberg received the Nobel Prize in Chemistry in 2006.
David Bushnell, Ken Westover and Roger Kornberg, Stanford University
View Media
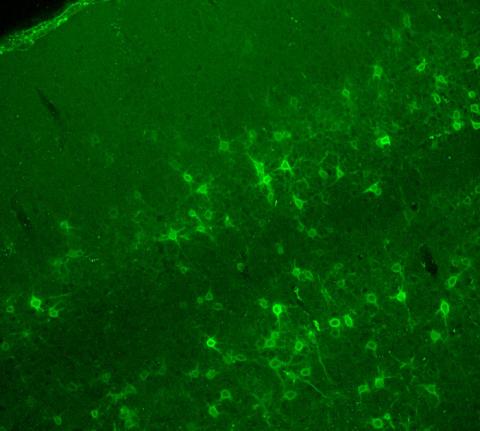
3742: Confocal microscopy of perineuronal nets in the brain 2
3742: Confocal microscopy of perineuronal nets in the brain 2
The photo shows a confocal microscopy image of perineuronal nets (PNNs), which are specialized extracellular matrix (ECM) structures in the brain. The PNN surrounds some nerve cells in brain regions including the cortex, hippocampus and thalamus. Researchers study the PNN to investigate their involvement stabilizing the extracellular environment and forming nets around nerve cells and synapses in the brain. Abnormalities in the PNNs have been linked to a variety of disorders, including epilepsy and schizophrenia, and they limit a process called neural plasticity in which new nerve connections are formed. To visualize the PNNs, researchers labeled them with Wisteria floribunda agglutinin (WFA)-fluorescein. Related to image 3741.
Tom Deerinck, National Center for Microscopy and Imaging Research (NCMIR)
View Media

2414: Pig trypsin (3)
2414: Pig trypsin (3)
Crystals of porcine trypsin protein created for X-ray crystallography, which can reveal detailed, three-dimensional protein structures.
Alex McPherson, University of California, Irvine
View Media
